Mice
Animal procedures were performed in accordance with national law (BGBLA 114 and Directive 522), European Directive 2010/63/EU and institutional guidelines for animal experimentation, and were approved by the Austrian Federal Ministry for Education, Science and Research (authorizations BMBWF-V/Sb: 2020-0.363.126, 2021-0.550.199, 2021-0.842.237, 2022-0.121.445 and 2023-0.930.355).
Mice were housed in groups of three to four under controlled laboratory conditions (12:12-h light–dark cycle with lights on at 07:00; 21 ± 1 °C; 55 ± 10% humidity) with food (pellets, 10 mm) and autoclaved water ad libitum. Mice were housed in commercially available individually ventilated cages made from polysulfone with a solid cage floor, dust-free bedding (woodchips) and nesting material.
For all experiments, male and female mice were used interchangeably to demonstrate the technology. Adult mice (aged typically two to three months, unless otherwise noted) were used as indicated with the following genotypes: C57BL/6J wild-type mice, Thy1-eGFP (STOCK Tg(Thy1-eGFP)MJrs/J mice, 007788, RRID:IMSR_JAX:007788 hemizygous) and haploinsufficient Hnrnpu+/− mice (deletion of one allele of Hnrnpu spanning axons 4 to 14, generated by crossing the HnrnpUWT/flox line (Hnrnpu<tm1.1Tman>/J, strain: 032187, RRID: IMSR_JAX:032187) with the CMV-CreCre/Cre line (B6.C-Tg(CMV-cre)1Cgn/J, strain: 006054, RRID: IMSR_JAX:006054).
Reagents
Chemicals and solutions are listed in Supplementary Tables 1 and 2.
Transcardial fixative perfusion
Solutions were prepared on the same day and kept at room temperature. Mice were first anaesthetized with isoflurane (1–2% (volume/volume; v/v) and then with ketamine (80–100 mg per kg body weight) and xylazine (10 mg per kg) intraperitoneally, combined with metamizol (200 mg per kg) subcutaneously for analgesia. After checking for deep anaesthesia by toe pinch, they were transcardially perfused at a flow rate of 7 ml min−1 using a peristaltic pump, first with room-temperature 1× phosphate-buffered saline (PBS) for 2 min and then with a solution (room temperature) containing 4% (weight/volume; w/v) paraformaldehyde (pH 7.4) and 10% acrylamide (AA, w/v) in 1× PBS for 6 min.
Brains were collected and post-fixed for 8–12 h at 4 °C with gentle agitation, using the same fixative solution.
Brain tissue processing
Brains were washed three times in cold (4 °C) 1× PBS for around 1 min each, with gentle agitation. They were sectioned coronally at a 50-µm thickness using a vibratome (Leica VT 1200 S). Where indicated, sections were prepared at a 300-µm thickness. Sections were placed in ice-cold 1× PBS supplemented with 100 mM glycine to quench PFA-reactive groups for 6–8 h at 4 °C and washed three times for around 1 min in 4 °C 1× PBS. They were stored at 4 °C in 1× PBS supplemented with 0.015% NaN3 for up to three months, typically.
Epoxide treatment
Brain sections were washed twice for 15 min each in 1× PBS at room temperature with gentle agitation. They were then washed twice for 15 min each in 100 mM sodium bicarbonate in milli-Q water, pH 8.0, at room temperature followed by incubation with 0.1% (w/v) TGE and 0.1% (w/v) GMA in 100 mM sodium bicarbonate in milli-Q water (pH 8.0) for 3 h at 37 °C with gentle agitation in a chemical hood. For 300-µm-thick slices, the TGE and GMA concentration was increased to 1% (w/v). Samples were then washed with 1× PBS for 1 h with gentle agitation at room temperature.
Pre-expansion immunolabelling for distortion analysis
Pre-expansion immunolabelling was performed only for distortion analysis measurements. All other immunolabellings were performed after expansion. Primary and secondary antibodies are listed in Supplementary Tables 3 and 4. Brain slices were permeabilized for 60–80 min with 0.2% Tween-20 in 1× PBS at room temperature with gentle agitation. Labelling with primary anti-GFP antibody was performed in 5% BSA and 0.2% Tween-20 in 1× PBS overnight at 4 °C with gentle agitation. Samples were then washed three to five times for around 30 min each in 1× PBS at room temperature with gentle agitation, followed by staining with secondary antibody overnight at 4 °C, in the same buffer as for the primary antibody labelling, with gentle agitation, followed by three to five washes for around 30 min each in 1× PBS with gentle agitation. Pre-expansion images were acquired on a spinning-disc confocal microscope after the first gelation step.
Preparation of hydrogel solutions
The compositions of hydrogels in LICONN are summarized in Supplementary Table 5. For preparation of the first hydrogel solution, AA and sodium acrylate (SA) were mixed in milli-Q water, vortexed and centrifuged at 4,500g for 5 min. Supernatant was transferred to a fresh 50-ml tube and supplemented with N,N′-methylenebisacrylamide (BIS) stock solution (prepared in milli-Q water) to a final BIS concentration of 0.075% w/v. After adjustment to the final volume, solutions were aliquoted in 2-ml tubes and stored at −20 °C, typically for up to one month. Solutions for the stabilizing and second expandable hydrogels were prepared similarly, usually freshly before experiments. We observed that the quality of sodium acrylate was crucial for successful expansion, mirrored in a batch dependency for this specific compound.
Polymerization of the first expandable hydrogel
In an ice-water bath, the first hydrogel monomer solution (Supplementary Table 5) was supplemented first with 0.001% 4-hydroxy-2,2,6,6-tetramethylpiperidin-1 (TEMPO, w/v) and then with 0.15% ammonium persulfate (APS, w/v) and 0.15% N,N,N′,N′-tetramethyl-ethylenediamine (TEMED, volume/volume; v/v) and vortexed. The following sample-handling steps were performed in an ice-water bath. Brain sections were pre-incubated with the hydrogel monomer solution for 30–45 min (75–90 min for 300-µm-thick slices) with gentle agitation in a 24-well plate. A gelation chamber was assembled from two cover glasses on bottom and top and #1 cover-glass strips on either side as spacers (for 300-µm sections: one #1 and one #1.5 cover-glass strip). Gelation was then performed at 37 °C in a humidified chamber for two hours. For optionally acquiring pre-expansion images, the sample was imaged on a spinning-disc confocal microscope (Oxford Instruments, Andor Dragonfly) after the first hour of gelation for around 15 min, and then further incubated at 37 °C for another approximately 45 min.
Disruption of mechanical cohesiveness, expansion and hydrogel neutralization
After gelation, the top coverslip was removed and the hydrogel–tissue hybrid was trimmed to the region of interest. Together with the bottom coverslip, it was then transferred to a 5-ml beaker with 2 ml denaturation buffer (200 mM SDS, 200 mM NaCl and 50 mM Tris-HCl in milli-Q water, pH adjusted to 9.0 with NaOH). The beaker was placed in a larger vessel containing pre-heated water and transferred to a water bath at 95 °C for 100 min. For 300-µm-thick samples, the denaturation time was extended to four hours, starting at 85 °C and ramping up to 95 °C within about 15 min.
For expansion, samples were placed in milli-Q water, with the water being changed every 20 min (approximately) until no further increase in gel size was observed.
We neutralized unreacted double bonds of the divinyl cross-linker (BIS) remaining after polymerization of the first hydrogel to ensure that it would not react with the following stabilizing and second expandable hydrogels. For this, we incubated with 0.2% APS plus 0.2% TEMED in milli-Q water for 2.5 h at 37 °C with gentle agitation, followed by one washing step with agitation in milli-Q water for 30 min at 37 °C and a similar washing step at room temperature.
Immunolabelling
Expanded hydrogels were again trimmed to the region of interest and incubated in 1× PBS for 10 min at room temperature, shrinking the gel in a 12-well plate. It was then incubated with primary antibodies (Supplementary Table 3) in 1× PBS at 4 °C overnight with gentle agitation in a 24-well plate, followed by at least three washes for one hour each in 1× PBS with gentle agitation at room temperature. Secondary antibody incubation (Supplementary Table 4) was also performed in 1× PBS at 4 °C overnight with gentle agitation, followed by at least three washes for one hour each in 1× PBS with gentle agitation at room temperature. Post-fixation was conducted for 15 min at room temperature with 4% PFA in 1× PBS, followed by quenching with 100 mM glycine in 1× PBS for 10 min at room temperature with gentle agitation and subsequent washing three times in 1× PBS for around 3 min each. Hydrogels were re-expanded in milli-Q water in a 12-well plate before further processing, and labelling was optionally evaluated by imaging.
During the LICONN procedure, cellular proteins are anchored to the hydrogel and epitopes can undergo denaturation and structural modification. We tested further primary antibodies by imaging directly after the immunolabelling; that is, without applying the second expansion step, as summarized in Supplementary Table 6. Note that not all of the antibodies that produce labelling in the first hydrogel may be retained throughout the entire sample preparation procedure. Dedicated optimization of heat and chemical denaturation and antibody incubation conditions may improve performance for individual antibodies. For somatostatin labelling, we applied a mixture of primary antibodies to increase the signal strength.
Polymerization of stabilizing hydrogel and neutralization
Expanded hydrogels were pre-incubated on ice-water for 3–3.5 h or 5 h for 50-µm-thick and 300-µm-thick slices, respectively, with the monomer solution of the stabilizing hydrogel (10% (w/v) AA, 0.025% (w/v) BIS, 0.05% (v/v) TEMED and 0.05% (w/v) APS in milli-Q water) with gentle agitation in a 12-well plate. After removing excess monomer solution, the hydrogel was sandwiched between a 22 × 22-mm2 coverslip placed on a microscopy slide and an 18-mm round coverslip on top of the hydrogel without spacers, and surrounded by monomer solution. Gelation was then performed for two hours at 37 °C in a pre-warmed (37 °C) humidified chamber. Gels were washed with milli-Q water for around 30–60 min at room temperature.
To neutralize remaining unreacted double bonds of the BIS cross-linker in the stabilizing hydrogel network and ensure that the hydrogel network did not react with the following second expandable hydrogel network, the hydrogel–tissue hybrid was incubated with 0.2% APS and 0.2% TEMED in milli-Q water for 2.5 h at 37 °C with gentle agitation, followed by washing with agitation in milli-Q water for 30 min at 37 °C and a second washing step at room temperature.
Polymerization of second expandable hydrogel
Stabilized and neutralized hydrogels were pre-incubated on ice-water with the monomer solution for the second expandable hydrogel (19% sodium acrylate, 10% AA, 0.025% (w/v) BIS, 0.05% TEMED and 0.05% APS in milli-Q water) in a 12-well plate for 3–3.5 h (50-µm slices), or 5 h (300-µm slices). After removing excess monomer solution, the hydrogel was again sandwiched between a 22 × 22-mm2 coverslip placed on a microscopy slide and an 18 × 18-mm2 coverslip on top of the hydrogel without spacers, and surrounded by monomer solution. Gelation was then performed for two hours at 37 °C in a pre-warmed (37 °C) humidified chamber. Hydrogels were washed with 1× PBS for around 30 min at room temperature.
Protein labelling
Pan-protein staining was performed with either 40 µM ATTO 488 NHS ester or 40 µM Alexa Fluor 488 NHS ester (Supplementary Table 7) in 1× PBS overnight at 4 °C with gentle agitation. Hydrogels were optionally washed three times for one hour in total with 1× PBS, and expanded for up to five hours before imaging in milli-Q water, with fluid exchange approximately every hour.
Mounting of expanded hydrogels for imaging
Before mounting for imaging, the region of interest was located with the spinning-disc confocal microscope in the expanded hydrogel after placing it onto a plastic culture dish with the bottom replaced with a 50-mm #1 cover-glass. For final imaging, the hydrogel was trimmed to the region of interest. Round coverslips (40 mm #1.5; Bioptechs) were rinsed with milli-Q water, covered with poly-l-lysine (PLL) and incubated at 37 °C for up to three hours. Coated coverslips were stored at 4 °C in 1× PBS for up to one week. Right before mounting, coverslips were washed with milli-Q water and placed in a home-built imaging chamber made from aluminium. The hydrogel was placed in the centre of the coverslip and further stabilized with two-component dental silicone (Twinsil extrahart, Picodent), and immersed in milli-Q water. We imaged for up to eight hours, typically.
Imaging using spinning-disc confocal microscopy
Imaging was performed on an Andor Dragonfly microscope based on a Nikon Ti2E inverted stand with motorized stage and an Andor Zyla 4.2 Megapixel sCMOS camera (2,048 × 2,048 pixels). Data were acquired using Andor Fusion software v. 2.2. Two pinhole disc patterns (25-µm and 40-µm hole diameter) and four continuous-wave excitation lasers (405 nm, 488 nm, 561 nm and 637 nm) were available. Imaging of non-expanded and expanded samples was performed with a 40× water-immersion objective (Nikon Apochromat LWD 40× lambda S, NA 1.15, water, working distance (WD) 0.6 mm), using the 40-µm disc pattern. For overview imaging (Fig. 5a and Supplementary Figs. 23, 24), a Nikon CFI P-Apochromat 20×, NA 0.95, WD 0.95-mm objective lens was used. The structural channel was imaged with a 488-nm excitation wavelength with a 521/38-nm detection bandpass filter. Typical exposure times were 110–150 ms and the laser power was set to 10–15% of available laser power. Voxels measured 150 × 150 nm2 laterally and the axial (z-direction) step size was chosen as 200 nm, 300 nm or 400 nm. The lateral field of view (FOV) was typically 307 × 307 µm2, corresponding to a biological tissue scale of around 20 × 20 µm2. The axial-imaging extent was typically chosen as 20–25 µm in biological tissue scale, limited by the 600-µm WD of the objective lens. For tiled measurements, the ‘field’ mode was used with 10% lateral overlap between individual subvolumes. For additional imaging of immunolabelling channels, colour channels (with excitation wavelengths of 488 nm (structural channel), 637 nm and 561 nm) were recorded sequentially in a frame-wise manner with exposure times of 120–200 ms per immunolabelling channel and typical laser power settings of 10–20%. For detection, 594/43-nm and 685/47-nm bandpass filters were used for the respective colour channels. Images for distortion analysis were acquired with a 150 × 150 × 200-nm3 voxel size with the 40×, NA 1.15 water-immersion objective lens.
Extension of imaging volumes in the z direction by block-face imaging and sectioning of expanded hydrogels
To seamlessly stitch imaging volumes not only laterally but also in the axial direction, we performed the following ‘lossless’ sectioning procedure, ensuring overlap of imaging volumes both laterally and axially. We expanded 300-µm-thick brain tissue sections and mounted them on PLL-coated coverslips for imaging as described above. We first imaged a multi-tile volume arranged on a 2D grid. We then removed the hydrogel and twinseal from the imaging chamber and glued the back surface (opposing the imaged layer) of the hydrogel onto the sample holder of a vibratome (Leica VT1200S, vibrating blade microtome) with superglue (Loctite 401, Henkel), gently flattening out the hydrogel by passing a soft plastic sheet over it. The vibratome chamber was partially filled with water, surrounding but not covering the hydrogel. We zeroed the vertical blade position on the hydrogel surface and lowered it to the desired cutting position, chosen to fall within the already imaged region (for example, cutting 320 µm below the hydrogel surface after imaging an axial range of 466-µm physical imaging range in the two-round measurement in Extended Data Fig. 10). Cutting was performed at 0.2 mm s−1 blade advancement. We then removed the hydrogel from the vibratome sample holder with the plastic sheet, mounted it for imaging with the vibratome-cut surface facing the PLL-coated coverslip and imaged a second layer of tiled, overlapping imaging volumes similarly to before. For the multi-round measurement, we iteratively repeated this procedure.
Troubleshooting
When establishing the procedure, we suggest that the protocol is adhered to closely, including details such as the precise chemicals and providers listed, the timing of steps, concentrations, temperatures at which individual steps are performed (for example, on ice-water versus at room temperature) and preparation of fresh solutions directly before experiments where indicated. On the basis of observations made during the development of the procedure (see for example Supplementary Figs. 1–5, 7–9 and Supplementary Table 8), we suggest checking the following points in the case of suboptimal results. Quality of transcardial fixative perfusion is crucial to achieve adequate tissue preservation, including, for example, cell shape. If tissue preservation is suboptimal, basic parameters such as perfusion speed and duration, and composition of solutions, should be checked. We anecdotally observed that when setting the pH of the perfusion solution, large pH variations should be avoided and it was preferable to carefully titrate in one direction until the final pH was reached. If the final expansion factor is lower than expected, over-fixation might be a factor (owing to an excessive duration of the post-fixation step, for example). Similarly, excessive protein anchoring might interfere with full expansion. Variation of cross-linker (BIS) concentration is also expected to have a direct effect on the expansion factor. For sodium acrylate, we observed a batch-to-batch variation in compound quality (for example, impurities), which could have an effect on the final expansion factor and overall outcome. For the other chemicals used, we did not make obvious similar observations. Shortening the time for expansion (for example, one hour instead of four to five hours for the final hydrogel) might also compromise the final expansion factor. Premature gelation of the second or third hydrogel might take place if the same reaction well is used for performing the preceding neutralizing step (removing unreacted double bonds) involving APS and TEMED, even if the well is washed. If the signal-to-noise ratio in the structural labelling is unsatisfactory, protein anchoring and labelling with NHS esters of fluorophores merit scrutiny. NHS esters are prone to hydrolysis. NHS esters of fluorophores should be dissolved only once needed in high-quality (water-free) dimethyl sulfoxide and aliquoted in single-use quantities (10-µl aliquots of 20 mM stock solution stored at −20 °C) to avoid freeze-thaw cycles, and should be used within two to three months. To monitor success at the various steps of the procedure during the set-up phase, it might be useful to perform imaging at various check-points, including after the first expansion step (applying NHS labelling), after immunolabelling and after full expansion.
Observing these guidelines produced highly consistent results. We followed the outcome of all transcardial fixative perfusions performed within a certain time period (n = 15 mice) from perfusion to expansion and final imaging. In total, the 15 perfusion experiments yielded 50 experimental runs that typically contained several technical replicates (hydrogel embedding and expansion experiments). In one out of 15 transcardial fixative perfusion experiments, the fixative perfusion itself failed, producing poor tissue preservation in the resulting samples, which is likely to be related to the suboptimal quality of the perfusion solution. In one technical replicate, we observed premature gelation of the stabilizing hydrogel, probably owing to remnants of APS and TEMED in the same well plate used during the neutralization step. The other replicates produced useful imaging data either for demonstrating various technical aspects (for example, determining the expansion factor, distortion analysis and testing of immunolabelling) or for the full LICONN procedure, as required. We selected a subset of these for further analysis and display, on the basis of visual inspection for factors such as the signal-to-noise ratio.
Data handling and format conversion
Raw imaging data were obtained in IMS format, as generated by the spinning-disc confocal microscope (Andor Dragonfly), which is a hierarchical data format with six resolution levels, from which we extracted the highest resolution level (level 0). Data conversion for downstream analysis was performed with custom Python scripts implemented in Python v.3.8 or higher, including the Imaris-ims-file-reader, zarr, webKnossos and tifffile packages. Custom scripts are available at https://github.com/danzllab/LICONN, with individual package versions specified in the respective configuration (.yml) files. Datasets are available through Neuroglancer (see Data availability section) or through the data repository of the Institute of Science and Technology Austria (ISTA)61 at https://research-explorer.ista.ac.at/record/18697. At various processing steps, conversion of file formats was necessary, and this was performed on ISTA’s high-performance computing cluster, using either a single node or SLURM (slurm-wlm 22.05.8) for task allocation, according to the resource requirement based on file size. We transferred files to cluster storage and converted from the respective source format to tiff, zarr, n5 or webKnossos (wk) format according to processing needs.
Image processing
Imaging data were processed using Fiji v.1.54f, including the CLAHE, BigWarp and BigData viewer plug-ins. For visualization, display ranges were adjusted, also accounting for a camera background signal of around 100. Intensity lookup tables were linear unless otherwise noted.
Overlay of immunolabellings with LICONN data was performed in GIMP v.2.10.34 after saving individual channels separately in RGB format in Fiji. The background in immunolabellings was set to transparent (alpha = black) and immunolabellings were overlaid with the structural channel. The output was saved in PNG format.
CLAHE (contrast-limited adaptive histogram equalization) was applied as indicated for individual figure panels, using either Fiji or a Python-based workflow as described below. In Fiji, first the display range was adjusted, and in the CLAHE plug-in the block size and histogram bin were set to the minimum and maximum values of the display range. The maximum slope was set to 3 and the standard (not fast) processing option was chosen. Three-dimensional renderings of cilia were done with Imaris v.9.3.
Distortion analysis and calculation of expansion factors
The analysis of distortions in the expansion procedure was adapted from previously published methods39,62, and the code was adapted from https://github.com/Yujie-S/Click-ExM_data_process_and_example. We analysed a total of 14 imaging volumes of around 20 × 20 µm2 laterally and 9–15 µm axially, recorded across 4 technical replicates in n = 3 Thy1-eGFP mice, exhibiting cytosolic expression of eGFP in a sparse subset of neurons. Imaging volumes corresponded to single tile measurements (no stitching or fusion of subvolumes). For distortion analysis, immunolabelling against eGFP was performed before subjecting the samples to the LICONN expansion procedure, and the eGFP channel was used for analysis. Pre-expansion images were acquired after around one hour of polymerization of the first hydrogel. Imaging volumes containing the same structures were acquired after expansion of the first hydrogel (before application of the stabilizing hydrogel) and after expansion of the second expandable hydrogel, with a spinning-disc confocal microscope and a 40×, NA 1.15 water-immersion objective lens, as in the LICONN imaging. This allowed us to assess distortions beyond the approximately 200-nm spatial scale. Using the BigWarp63 plug-in in Fiji, we first manually placed around 25 landmarks at corresponding neuronal features in maximum intensity projections of the respective pre- and post-expansion imaging volumes. Using BigWarp, we then applied either a similarity transformation (including isotropic scaling, translation and rotation) or an affine transformation (including scaling, translation, rotation and shearing) to the pre-expansion images to best match landmarks in pre- and post-expansion images. Expansion factors were extracted as the linear scale factor in the similarity transformations. We smoothed pre- and post-expansion projection images with Gaussian filters of different widths to account for the different resolution in the respective images. For individual expansion steps, σ was set to 1 and 6 pixels for pre- and post-expansion images, respectively. For total (iterative) expansion, σ was set to 1 and 16 for pre- and post-expansion images, respectively, resulting in a comparable appearance as judged by visual inspection. For one of the datasets, the intensity range was slightly saturated at the main dendrite branch in the pre-expansion image. We now used the imregdemons.m function in MATLAB (v.R2022b, MathWorks) to compute distortion vector fields62,64 from the aligned pre- and post-expansion images. For this, we used the images before the respective expansion step as a reference and evaluated distortions arising from that expansion step. For display purposes, we used the quiver function in MATLAB with vector scaling set to 1.5. We then computed and plotted distortions (measurement error) for different measurement lengths, as previously described16. For this, we generated binary masks of neuronal structures of interest by thresholding pre- and post-expansion images, and then combining, dilating and eroding the resulting masks using parameters as judged by visual inspection. We then randomly sampled 2 × 105 pairs of points restricted to the masked area of interest. These pairs of points defined a set of vectors in the pre-expansion image. We then found a corresponding set of transformed vectors by applying the distortion field generated with imregdemons.m to the respective pairs of points. For each measurement length, we then computed the measurement errors as lengths of the difference vectors between the pre-expansion and transformed vectors combined from three or four different datasets within one experiment. For plotting, measurement errors at a given measurement length were averaged and the mean was plotted together with the standard deviation.
From these measurement errors, the root mean square (RMS) error was calculated across different measurement scales. Finally, the mean and standard deviation of RMS errors across the n = 4 technical replicates were plotted for individual expansion cycles and transformations. For display purposes, we smoothed the measurement error curve with a one-dimensional median filter.
Stitching and fusion of imaging tiles
We extended SOFIMA (https://github.com/google-research/sofima, git hash 64d5c7c) to support seamless stitching of 3D tiles. In brief, we processed every volumetric tile with CLAHE, applied independently to every in-plane (xy) section of the tile. The tiles had an in-plane size of 2,048 × 2,048 voxel2 and an axial extent varying depending on the experiment. We laid out the tiles on a 2D grid according to imaging metadata, and used a coarse positioning step to find their initial location in the coordinate system of the output volume. This was followed by fine alignment to elastically deform them to obtain seamless matches between adjacent tiles and form 3D sections, around 20–30-μm thick. The tiles were then rendered with linear-distance-weighted blending of overlapping image content.
For the coarse positioning step, we extracted a small (50-voxel)3 cube of image content from the ‘preceding’ tile of every adjacent tile pair, and used cross-correlation to identify the corresponding image content in the ‘following’ tile, forming an offset vector between the tiles. The tiles were then modelled as unit masses connected by springs of resting lengths equal to the computed offsets, and the system was allowed to relax, with the relaxed state determining the tile positions at the beginning of the fine alignment step.
For fine alignment, we modelled every tile as an elastic mesh of unit masses placed on a regular grid with a 3D stride of 20 voxels and nearest neighbour masses connected with Hookean springs. Coarse positioning resulted in the neighbouring tiles partially overlapping with each other, and for any mass located in the overlapping image region, we again used cross-correlation to identify the position within the neighbouring tile of a an (80 voxel)3 image patch centred on the mass (PATCH_SIZE=80). These formed point correspondences between the tiles, which we modelled as 0-length Hookean springs, and allowed the whole system to relax. The relaxed set of tile meshes was used to render the aligned volume.
For datasets comprising more than one thick section (Extended Data Fig. 10), we first stitched tiles into thick sections as described above, and then aligned them to each other sequentially. First, we manually identified the approximate amount of overlap in the axial dimension (typically 100–200 voxels) for adjacent sections, and computed a flow field from the middle plane of the overlap subvolume of the following section to the overlap subvolume of the preceding section, using a patch size of (80, 80, 80) and a stride of (40, 40, 40) voxels in the following section, and a patch size of (160, 160, overlap depth) in the preceding section.
We then modelled the following section as a 3D spring mesh, and allowed it to relax according to the flow field. The relaxed mesh was used to render the following section in alignment with the preceding one. We tracked the position of the middle plane of the following section for which the flow field was computed as the thick section was warped, and when fusing it with the preceding section, retained voxels from the preceding section above (lower axial coordinates) this warped plane, and used voxels from the following section below it. This is similar to the ‘fine alignment’ of individual tiles, and the overall strategy is a direct 3D analogue of how alignment of 2D sections is done with SOFIMA for volume EM datasets.
Details of processing were as follows. In all cases, we based stitching and fusion on the structural imaging channel. We first transferred individual raw IMS files (uint16 format) to cluster storage and extracted the highest resolution level (level 0). We then used a BigStitcher-based Fiji macro to concatenate the data and convert to n5 format. We then applied CLAHE tile-by-tile with Python. Data handling was done with the TensorStore library (https://google.github.io/tensorstore/, v.0.1.33). We used the numpy.clip function to clip the intensity range plane by plane typically between 120 and 350, determined by visual inspection. For CLAHE, we used skimage.exposure.equalize_adapthist to apply histogram equalization, with a clip limit of 0.03. We multiplied the output (ranging from 0 to 1) with 255 and saved as uint8 format.
In the rigid, coarse alignment step, we determined the relative arrangement of tile pairs inputting the known tile layout (arrangement of tiles in the image acquisition). We set the QUERY_R_ORTHO and QUERY_R_OVERLAP parameters to 25. We set the minimum tile overlap (QUERY_OVERLAP_OFFSET) to 60 (100 for the dataset in Fig. 2a) and the size of the search area within tiles (SEARCH_OVERLAP, SEARCH_R_ORTHO) to 300 (400 and 600, respectively, for the dataset in Fig. 2a). The second, fine-stitching, step was used to warp tiles and achieve a smooth transition at tile borders, using a stride of (20, 20, 20) voxels. This step outputs a mesh that ensures a globally consistent warping and relative arrangement of individual tiles. In the last step, we applied the obtained mesh to either the CLAHE-modified structural imaging data for downstream (automated) segmentation or skeletonization, or the raw structural imaging data, as well as to immunolabelling channels, resulting in the respective fused imaging volumes in n5 format. We performed these processing steps on ISTA’s high-performance computing cluster and/or, in the case of the measurement in Fig. 2a and Extended Data Fig. 10, on Google’s high-performance computing infrastructure, requesting resources according to the processing task. Typically, for CLAHE, we requested 16 CPUs (Intel Xeon CPU E5–2680 v4, 2.40 GHz or similar) and 150 GB of RAM. For coarse stitching, we typically requested 3 GPUs (GeForce RTX 2080 Ti or similar), 32 CPUs and 100 GB or RAM. Fine stitching was done with 1 GPU, 16 CPUs and up to 970 GB of RAM. Rendering was typically done requesting 32 CPUs and up to 700 GB of RAM.
Manual segmentation and proofreading
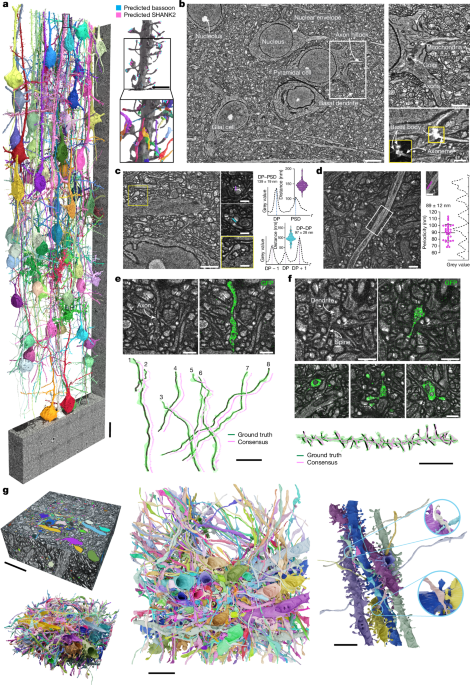
For manual segmentation, we used VAST v.1.4.0 (downloaded from https://lichtman.rc.fas.harvard.edu/vast/) with the data in eight-bit format. Segmentation of the data in Fig. 1g was originally performed by one person, and the resulting segmentation was used for training. For visualization in Fig. 1g, segmentation was partially proofread by another person using VAST v.1.4.1.
Skeletonization
We used webKnossos v.22.05.1 for manual skeletonization, running on a local computer with 2× AMD EPIC MILAN 75F3 processor, 32-core 32C/64T, 2.95 GHz with 512 GB of memory, 2 TB NVMe-SSD and an additional 27 TB NVMe-SSD, as well as 4 NVIDIA A6000 GPUs. To reduce required memory resources, we converted data to uint8 format when converting to the webKnossos (wk) format. Before conversion, we clipped the intensity range according to visual inspection, to account for intensity outliers and avoid signal overflow. For convenient exploration of the datasets across spatial scales, we made use of ten precomputed resolution levels, using the webKnossos downsample function. Typically, raw data rather than CLAHE-processed data were used for tracing. When comparing automated segmentation of the volume in Fig. 2a with manually generated skeletons, we checked sites of discrepancy and corrected skeletons as appropriate.
Generation of GFP-based ground truth
For testing traceability by comparison with an independently generated ground truth, we compared manual tracings of neurites in the structural LICONN channel with their structure revealed by cytosolic expression of eGFP in Thy1-EGFP mice. The LICONN structural channel and the eGFP channel were recorded at a similar spatial resolution after tissue expansion with a spinning-disc confocal microscope. The structural channel was recorded in the 488-nm excitation channel, whereas eGFP was detected by post-expansion immunolabelling as described above with imaging at 561 nm excitation. Voxel sizes were either 150 × 150 × 200 nm3 or 150 × 150 × 300 nm3, with the largest step size referring to the z direction. Data were converted to wk format for tracing. Using the webKnossos access permissions feature, we assigned permissions to ensure blinding as required. For generating ‘ground-truth’ skeletons of eGFP-expressing neurons, both the structural and the eGFP channels were loaded and considered jointly. In addition, one seed point was placed in each eGFP-expressing neuronal structure. Two novice human tracers were trained in a non-blinded manner with both the eGFP and the structural LICONN channels of four datasets (single volumetric imaging tiles, around 19 × 19 × 19 µm3), containing, in total, two eGFP-expressing axons and two eGFP-expressing dendrite stretches. These datasets were excluded from further analysis. After training, the LICONN channel together with the seed points were provided to the two human annotators blinded to the eGFP channel who independently traced the marked structures based on the structural channel.
For dendritic spines, annotators were asked to indicate these as short branches diverging from the main dendrite trunk. The annotators were then asked to compare the individual skeletons and generate a ‘consensus’ skeleton for each of the traced structures. Human consensus skeletons were then overlaid with and compared to the eGFP-based ground truth using webKnossos.
Neuronal instance segmentation
We used FFNs (available at https://github.com/google/ffn, git hash 12d680e)42 to automatically segment the datasets. We used the original convolutional stack architecture with a 33 × 33 × 33-voxel FOV, a step size of 8 voxels and the network depth extended to 20 residual modules. The models were applied to data with 19.4 × 19.4 × 25.9-nm3 voxel size (in original tissue scale). For this, CLAHE-processed volumetric imaging data were 2× downsampled laterally relative to the imaging voxel size using volume averaging in Python. Axially, datasets with a 200-nm axial-imaging step size (effective voxel size 9.7 × 9.7 ×13.0 nm3, obtained by scaling with an exF of 15.44; Fig. 2a) were also downsampled twofold in the z direction, whereas datasets with a 400-nm axial step size (effective voxel size 9.7 × 9.7 × 25.9 nm3; Fig. 1a) were not downsampled in the z direction. Training was performed with a batch size of 128 and a learning rate of 10−4 using the AdamW65 optimizer with 32 NVIDIA V100 GPUs, for up to 30 days, which corresponded to about 8.8 million training steps. A Jupyter notebook for performing FFN inference on LICONN data is available at https://github.com/google/ffn/blob/master/notebooks/ (git hash: 12d680e).
We used an iterative approach to collect the volumetric ground-truth annotations for training the FFNs. First, 162 neurite fragments covering 110 Mvx (megavoxels, at full imaging resolution) were manually annotated as described above, using VAST in the dataset shown in Fig. 1g (single tile). Model 1 trained with these data on full-resolution imaging data was used to segment the multi-tile volume in Fig. 2a. We then manually agglomerated 103 neurite fragments covering 447 Mvx (voxel number referring to full imaging resolution) within that segmentation (for a description of manual agglomeration, see below). We used this to train another FFN model (model 2), using both the newly collected data and the pre-existing manually painted annotations for training with voxels 2× downsampled in every direction (19.4 × 19.4 × 25.9-nm3 effective voxel size). Within the segmentation results of model 2, we manually proofread a larger set of axons (540 axons, 920 Mvx at full imaging resolution, 115 Mvx downsampled, 17.8-mm cumulative path length) and dendrites (314 dendrites, 3,344 Mvx at full imaging resolution, 418 Mvx downsampled, 23.8-mm cumulative path length), and used these data to train the final (model 3) FFN model that was used to produce all the automated neurite segmentations in the manuscript.
We followed a segmentation strategy optimized for first reducing the number of segments affected by merge errors (‘base segmentation’) and then agglomerating these mostly merge-free supervoxels into larger neurites, as described in the following sections. For the base segmentation, we applied the seed-order oversegmentation consensus procedure42, followed by oversegmentation consensus with two more segmentations generated with alternative FFN check-points (snapshots of weights saved at regular time intervals during training), which were manually selected for having a small number of wrong merges. This reduced the number of falsely merged supervoxels in the segments evaluated with manually generated skeletons in the volume in Fig. 2a to zero, whereas the overall volume contained at least five mergers. For determining edge accuracy42, a skeleton edge was counted as ‘accurate’ when both nodes were labelled identically and the label did not correspond to a segment overlapping more than one skeleton.
We automatically skeletonized the base segmentation for downstream use in agglomeration and proofreading using the TEASAR algorithm66 (https://github.com/seung-lab/kimimaro, version b390a9abdde60ec81472f1901e7980a962ecd847) and heuristically identified all nodes of degree 1 in the resulting skeleton graphs as ‘end-points’.
Semantic segmentation
To automatically classify segments into distinct subclasses—in particular, axons and dendrites—we developed a semantic segmentation pipeline.
For this, an experimenter classified manually agglomerated objects in a segmentation of a 512 × 512 × 334-voxel subvolume (at the downsampled 19.4 × 19.4 × 25.9-nm3 effective voxel size) of the dataset in Fig. 2a into one of five classes: axons (331 segments, 15 Mvx), dendrites (36 segments, 11.1 Mvx), myelinated axons (6 segments, 1.1 Mvx), oligodendrocytes (2 segments, 0.15 Mvx) and (astro)glia (47 fragments, 1.3 Mvx). We used these annotations to train a neural network model to perform semantic segmentation (voxel-wise multi-class prediction) of the structural LICONN data. The neural network used the same residual 3D convolution stack architecture as the FFN, with a depth of eight residual modules and a FOV of 33 × 33 × 33 voxels, but with the convolutions operating in ‘valid’ mode, and the residual summation discarding context at the boundary of the larger-sized argument. The model was trained with asynchronous stochastic gradient descent at a learning rate of 10−4, using 16 NVIDIA V100 GPUs and a batch size of 16. Training lasted approximately 38 h, during which 40.7 million steps were executed. Training points from all five classes were sampled at equal frequencies.
We used the trained model to generate a semantic segmentation of the entire volume in Fig. 2a and associate every voxel with the class predicted with the highest probability. These voxel-wise classifications were then aggregated over all voxels of every instance segment, and the class predicted most frequently was associated with that segment.
Segment agglomeration
The instance segmentation pipeline described above is optimized to make it unlikely that an individual super-voxel incorrectly covers more than one neurite. This happens at the cost of an increased number of split errors; that is, assigning multiple smaller segments to the same neurite.
To mitigate this, we applied a multistep agglomeration procedure in which the FFN model was used to evaluate whether a pair of geometrically proximal segments should be merged together42 and how a segment could be further extended from a heuristically detected end-point in the skeletons generated from the base segmentation9. In the first step, we used the ‘relaxed acceptance criterion’9. In the second step, we merged two segments when the end-point-seeded FFN inference recovered 90% of the voxels of both segments within a (200 voxel)3 subvolume centred at the end-point.
We further trained two FFN models with a 3D U-net architecture and a 1283-voxel FOV and without a FOV movement policy. We used the same training data as for the main instance segmentation model, but split the data into two non-overlapping sets, so that one model was trained exclusively on axons, and the other exclusively on dendrites. We applied the axon model to end-points of all axon segments. Within the prediction results for every end-point, we determined the segment S with the highest recovery fraction (other than the segment E containing the end-point) and agglomerated S and E if that fraction was at least 50%, and if S had an end-point within 100 voxels of the end-point of E. We applied the dendrite model to all skeleton nodes of dendritic trunks (skeletons generated from the base segmentation), taking any dendritic segment of at least 100,000 voxels to be a trunk. For every evaluated point, we collected any putative spine (dendritic segment of less than 100,000 voxels) associated with at least a 50% recovery fraction. We then agglomerated all spines to the trunk for which it had the highest recovery fraction.
The agglomeration procedure resulted in a graph with 516,437 edges, in which each edge represents one pair of automatically joined base segments.
Agglomeration graph filtering
The semantic segmentation model allowed us to associate per-class voxel counts with all base and agglomerated segments. We used these classifications to filter the agglomeration graph similarly to previous work9. Specifically, we sorted the edge list in decreasing order of the associated scores, and discarded any edge that would cause a merge between inconsistent classes. The class combinations we considered were: (astro)glia versus axons versus dendrites versus oligodendrocytes; (astro)glia and oligodendrocytes versus others; and (astro)glia and oligodendrocytes versus axons versus dendrites. An (agglomerated) segment was considered to belong to one of these composite classes when it had a total size of at least 100,000 voxels, and at least 75% of its voxels were classified as the target class. This criterion was relaxed to 10,000 voxels and 50%, respectively, for the end-point agglomeration step, in which we also disallowed merging together objects larger than 100,000 voxels.
Manual proofreading of neuron segmentations
The automated segmentation of the dataset obtained as a result of the agglomeration procedure was expected to have remaining split and merge errors, which we decided to correct in a structured manual proofreading workflow, leading to a marked increase in proofreading speed. Focusing on topological split and merge errors rather than painting voxels, the workflow was implemented as a Python script driving the Neuroglancer viewer67 (https://github.com/google/neuroglancer, c466b24), within which an expert annotator interactively inspected segments and the structural channel images in both 2D cross-sections and as 3D mesh rendering. The tool had features for automatically moving the 3D cursor to predetermined locations (such as end-points of segments), marking segments for future view and directly editing the agglomeration graph by adding or removing edges.
Using the results of the semantic segmentation to classify all agglomerated segments, we organized the proofreading process as follows:
-
1.
All axon segments (n = 2,674) of at least 10,000 voxels and touching a single face of the bounding box of the volume were inspected for splits and manually connected to appropriate partner segments where necessary. The tool was configured to automatically navigate to heuristically detected end-points of the axon segments.
-
2.
All axons within the volume (n = 18,667) were reviewed for shape implausibilities resulting from putative merge errors, which were then inspected and corrected.
-
3.
Dendritic spines not connected to a trunk (n = 2,782) were reviewed and connected to a trunk when one could be unambiguously identified.
-
4.
All dendritic branches (n = 1,643) were reviewed for incorrectly associated spines, which were separated, and subsequently manually connected to the correct trunk.
Synapse detection through immunolabelling
To automatically detect pre- and postsynaptic sites in volumetric datasets comprising the LICONN structural channel and immunolabelling channels for bassoon and SHANK2, respectively, we loaded the multicolour image stacks in tiff format. We converted image stacks for molecular signals to 32-bit floating numbers and adjusted the contrast to a minimum of 101 to account for camera background, and the maximum to the 99.5th and 99th percentile for the dataset in Fig. 4l and Supplementary Fig. 19g–i (300-nm z-step size) and 99.95th and 99th percentile for the datasets in Figs. 3g,h and 4p (200-nm or 400-nm z-step size) for bassoon and SHANK2, respectively. We then applied background subtraction by applying Gaussian filters of two different widths using scipy.ndimage.gaussian_filter, with σ = 5 voxels and σ = 11 voxels, corresponding to signal and background, respectively, for bassoon (σ = 6 voxels and σ = 10 voxels for datasets with 200-nm z-step size) and σ = 4 voxels and σ = 11 voxels (σ = 6 voxels and σ = 12 voxels for datasets with 200-nm and 400-nm z-step size) for SHANK2. Background was subtracted from signal and negative values were set to zero. We then applied Otsu thresholding and used the skimage.measure.label function for converting the resulting binary mask into instance segmentations. We then applied the regionprops function to extract the maximum intensity value for the underlying LICONN channel for each of the 3D regions defined by the individual segments. The list of maximum intensity values had a bimodal distribution, which we used to distinguish between synaptic regions (high intensity in structural channel) and unspecific immunolabelling (low intensity in structural channel). To separate the two populations, we again used Otsu thresholding. To remove further non-specific detections, we performed additional size filtering. Segments that occupied less than 60 voxels were removed, as well as objects that spanned fewer than a certain number of planes in the z direction (300-nm z-step size: <13; 200-nm z-step size: <14 for bassoon and <12 for SHANK2; 400-nm z-step size: <12 for SHANK2). Listed parameters were determined from a grid search on a separate validation dataset or a subvolume of the analysed dataset for each of the z-step sizes, choosing the parameter set that resulted in the highest F1 score.
We finally converted the individual segments into point annotations, using the regionprops function to find the weighted centroid for each segment based on the underlying LICONN data.
For detecting gephyrin-positive post-synapses, we applied a simplified algorithm involving background removal, Otsu thresholding, morphological opening and size filtering, and manually proofread the output.
Validation of immunostaining-based pre- and post-synapse detection
For validating automated synapse detection, a human annotator created ground-truth synapse annotation in a dataset comprising the LICONN structural data, bassoon immunolabelling and SHANK2 immunolabelling. For each synapse, they generated a ‘skeleton’ with the source point in the pre-synapse and the target point in the post-synapse, near the respective centres of the molecular signals, using webKnossos v.22.05.1, after conversion of the datasets to wk format. Bassoon signals without corresponding SHANK2 signals, which are likely, in large part, to correspond to inhibitory synapses, were annotated as single source points. Similarly, at volume edges, instances of SHANK2 signals without corresponding bassoon signals occurred, which were annotated as single target points. The human annotator also indicated synaptic connections that lacked SHANK2 signal but showed a clear PSD in the structural channel as excitatory post-synapse. The special cases of a single post-synapse connected to more than one pre-synapse or a single pre-synapse connected to multiple post-synapses were handled by text comments. In total, three datasets were annotated. One test dataset from the hippocampus had a 300-nm z-step size and comprised 700 × 700 × 200 voxels of size 9.7 × 9.7 × 19.4 nm3. This dataset contained 261 pre-synapses and 247 post-synapses, forming 242 full synapses (with one-to-many connections contributing a count of 1 with each edge), with 25 occurrences of unpaired pre-synapses and 7 unpaired post-synapses. The second test dataset from the hippocampus had a 200-nm z-step size and comprised 1,000 × 1,000 × 600 voxels of size 9.7 × 9.7 × 13.0 nm3. This dataset contained 1,116 pre-synapses and 1,084 post-synapses, forming 1,059 full synapses (with one-to-many connections contributing a count of 1 with each edge), with 57 occurrences of unpaired pre-synapses and 25 unpaired post-synapses. The third test dataset was from cortex, had a 300-nm z-step size and comprised 1,322 × 1,322 × 170 voxels of size 9.7 × 9.7 × 19.4 nm3. This dataset contained 306 pre-synapses and 262 post-synapses, forming 261 full synapses (with one-to-many connections contributing a count of 1 with each edge), with 48 occurrences of unpaired pre-synapses and one unpaired post-synapse. We generated further validation datasets for parameter tuning, which were not included in the test data.
To compare the automatically generated point annotations of bassoon or SHANK2 signals with the corresponding human ground-truth point annotations, we performed spatial matching of the respective point annotations. For this, we defined a cost matrix with entry Cij corresponding to the Euclidean distance between automatically generated point annotation i and manually generated point annotation j. We defined a matching limit rmatching; that is, an upper distance beyond which matching was not allowed, and set it to rmatching = 30 voxels after stepping this parameter in a test dataset. For Cij > rmatching, we set this matrix entry to an arbitrary large number. We now used the scipy.optimize.linear_sum_assignment function to solve the linear assignment problem of finding corresponding points; that is, the single-point to single-point matches with minimum Euclidean distances. Pairs matching between automatically and manually generated annotations were classified as true-positive automated detections. Automated detections without corresponding ground-truth element within the matching radius rmatching were classified as false-positive detections. Ground-truth detections without a corresponding match in the automated detection were classified as false-negative detections.
Association to full synapses
To connect corresponding presynaptic sites automatically identified by bassoon immunolabelling with postsynaptic sites automatically identified by SHANK2 immunolabelling, we first defined a cost matrix with entries Sij corresponding to the Euclidean distances between presynaptic annotations j and postsynaptic annotations i. We then defined a binary matching matrix, Mij, with Mij = 1 indicating a connection between the respective pre- and postsynaptic sites (else 0).
Solving this as a linear assignment problem as above would result exclusively in 1:1 matches, which would not capture cases in which one pre-synapse is connected to multiple post-synapses or vice versa. We therefore developed a pipeline that gave priority to the spatially closest occurrences of bassoon and SHANK2 during the presynaptic–postsynaptic pairing procedure but that also allowed for one-to-many connections.
In a first pass, we paired every postsynaptic site with the closest presynaptic sites; that is, we identified the smallest Sij within each row of the cost matrix and set the corresponding Mij to 1 if Sij was below a maximum matching distance of dmatching = 30 voxels. This value was taken from the distance distribution between manually generated pre- and postsynaptic annotations, choosing a value close to the observed maximum distance of 31 voxels.
Hence, each post-synapse was at most associated with one pre-synapse, whereas each pre-synapse could be associated to zero, one or more post-synapses. In a second pass, we now exclusively considered those bassoon signals that were left unpaired in the previous run (‘floating’ pre-synapses); that is, bassoon occurrences j that did not have Mij = 1 after the first pass. For these, we found the postsynaptic entry i with the lowest distance and set the corresponding Mij to 1 if this distance was below the matching limit dmatching. This identified the previously unpaired pre-synapses that had SHANK2 signal in close spatial proximity; that is, within the matching limit.
First associating the closest pre- and postsynaptic sites in this way did not allow additional connections between pre- and postsynaptic detections that were already assembled into a synaptic connection in a more closely matching pair, which would occur if we simply matched all pre- and postsynaptic occurrences within the maximum matching distance dmatching.
Validation of synapse assembly
We classified automated detections of synaptic connections (that is, a connection between an individual presynaptic annotation and an individual postsynaptic annotation) as true positive if the following criteria were fulfilled: (i) true-positive detection of a bassoon occurrence; (ii) true-positive detection of a SHANK2 occurrence; and (iii) an automatically detected connection between these two that was also present in the manually annotated connections. We classified automated detections of synaptic connections as false positive if (i) a presynaptic and a postsynaptic detection were automatically connected but were not connected in the manual annotation or (ii) a false-positive molecule detection (bassoon or SHANK2 prediction) was assigned a synaptic connection. We classified detections of synaptic connections as false negative if a manually annotated synaptic connection was not present in the automated detection of synaptic connections. A typical scenario for this was a missed automated detection of pre- or postsynaptic sites, which can also happen if two neighbouring bassoon occurrences (or SHANK2 occurrences) are blurred into each other during the smoothing step and thus undersegmented.
For determining F1 scores, we revised false-negative detections that were close to the border of the analysed imaging volume in the z direction, because potential true-positive detections near the border might have been removed during the z-size filtering step. For this, we automatically generated two lists of detections: one that included objects spanning fewer than the specified number of planes in the z direction and a second in which these structures were removed. Further true-positive detections were then defined according to the following rules: (i) the detection was true positive in the first list and false negative in the second list and (ii) the detection was located within the first ten or last ten imaging planes of the volume.
Automated post-processing of synapse detections
To account for those cases in which a particular synapse was not retained during one of the processing steps for automated synapse detection, we implemented an automated post-processing step for the respective test datasets (Supplementary Fig. 19). Such cases included failed detection in cases in which the synaptic immunolabelling was either weak or absent at a particular synapse and therefore did not meet the cut-off threshold (for example, owing to lack of SHANK2 expression), or the sampling of the structural LICONN feature did not pass the global threshold, or size filtering. Here, we identified unpaired pre- and postsynaptic detections, defined a local search area around those and checked whether bright structural features were present in the LICONN channel around those unpaired detections. For cases in which a prominent PSD or bright presynaptic feature was present, we classified them as additional pre- or postsynaptic detections, and added them to the respective lists of pre- and post-synapses, including them in the presynaptic–postsynaptic matching algorithm.
In brief, we checked individual rows and columns of the matching matrix Mij that only contained zeros, corresponding to unmatched post- and pre-synapses, respectively. We then defined a 100 × 100 × 100-voxel3 bounding box around those locations (or smaller at imaging volume edges). We then applied intensity rescaling to the structural LICONN channel between p1 and p99.95, where p1 refers to the first percentile and p99.95 to the 99.95th percentile. We then removed background using Gaussian filters of two different widths at σ = 2 voxels and σ = 10 voxels, subtraction and clipping of negative values to zero. We then performed thresholding at thr = p95 + 0.4 × (p100 − p95), with p95 and p100 being the 95th and 100th percentile, respectively. For the dataset in Supplementary Fig. 19g–i, a factor of 0.5 instead of 0.4 was used. We then removed previously detected pre- and post-synapses by multiplying the resulting mask with a mask corresponding to an inverted semantic segmentation of the previously detected pre- and post-synapses. We then did a binary opening (eroding and then dilating) with a ball of two-voxel radius. We then performed instance segmentation with skimage.measure.label, removed structures smaller than 20 voxels and determined the weighted centroid on the intensity distribution in the underlying structural LICONN channel. We classified the resulting objects as a new pre- or postsynaptic detection when their Euclidian distance from the originally unpaired pre- or postsynaptic site was smaller than the matching limit rmatching. We used these to update the list of pre- and postsynaptic detections used for the association to full synapses.
Deep-learning-based synapse detection
We devised a strategy for identifying the locations of pre-synapses and post-synapses from the structural imaging channel alone. We used deep-learning-based prediction of synapse location with an approach that did not require human input for the generation of training data. For this, we implemented a U-net architecture for image translation to predict the locations of synaptic molecules from structural data, using measured molecule locations as input to the training. We then converted the predicted molecule locations to automatically generated synapse detections, for this step using a pipeline that was similar to the immunolabelling-based pipeline above. We generated two independent models for predicting the location of the presynaptic marker bassoon and for predicting the excitatory-synapse-specific postsynaptic marker SHANK2.
Generation of training data
We converted 3D super-resolved measurements of synaptic-molecule locations (based on specific immunolabelling), paired with the structural imaging channel, to training data with the following workflow. We first converted TIFF images to 32-bit floating format and split them into individual datasets for the structural and each of the immunolabelling channels, maintaining the original voxel size. We then implemented a preprocessing pipeline to automatically remove unspecific immunolabelling signals, similar to the approach taken for the immunolabelling-based synapse detection described above. However, unlike in that pipeline (in which we performed a strong smoothing step to avoid oversegmentation of the spatially structured bassoon and SHANK2 distributions), we here preserved the spatial distribution of the bassoon and SHANK2 signals more closely, allowing the deep-learning network to recapitulate the molecule arrangement in the respective synaptic compartments, including the lattice-like arrangement of bassoon, paralleled by high-intensity (protein-rich) structural features.
We rescaled intensity using the skimage.exposure.rescale_intensity function, setting the minimum to 101 and the maximum to the 99.995th percentile. We then removed background by applying isotropic Gaussian filters of two different widths using scipy.ndimage.gaussian_filter, with σ = 0.5 voxels and σ = 10 voxels, corresponding to signal and background, respectively, subtracting background from signal and clipping negative values to 0. We applied Otsu thresholding to generate a binary mask and used the skimage.measure.label function to derive an instance segmentation. We then sampled the maximum intensity value of the underlying structural data using the skimage.measure.regionprops function. We applied Otsu thresholding to distinguish segments with underlying high-intensity LICONN features, which we classified as specific immunolabellings, and without high-intensity features, which we classified as non-specific immunolabellings. We next removed the latter and converted the remaining segments back to an overall binary mask. To approximate the intensity distribution of the immunolabellings, we blurred the resulting mask with a 3D Gaussian of σ = 1 voxel.
For the structural channel, we applied simple intensity normalization to image pixel values im (im’ = (im − min)/(max − min)), where min is the 0th percentile and max is the 100th percentile of the intensity distribution. We converted to 8-bit format by multiplying the normalized data by 255 and clipping the intensity range between 0 and 255, followed by type casting.
We saved the paired immunolabelling and structural data in zarr format, allowing resource-efficient handling of large datasets.
Network architecture and training
We implemented deep-learning pipelines with Pytorch v.1.12.1 (https://pytorch.org) and used the Gunpowder framework v.1.2.2 (https://github.com/funkelab/gunpowder) to implement our data loading, augmentation, training and prediction pipeline that conveniently allowed processing of big datasets. For predicting molecule location, we used the 3D U-net architecture from a previous study46. We used Adam’s optimizer (torch.optim.Adam) with a 10−4 learning rate. We used the torch.nn.MSEloss function to implement mean squared error as a loss function. The batch size was set to 8 and the training input size was 64 × 128 × 128 voxels, with the smallest number referring to the z direction.
We performed network training on ISTA’s high-performance computing cluster, using SLURM for task allocation. We typically requested 8 CPUs, graphics acceleration by 1 GPU (NVIDIA GeForce) and 700 GB of RAM.
We trained on datasets with a 300-nm z-step size (6 individual tiles, total volume of 16,699 µm3, containing around 16,250 excitatory synapses) and applied the resulting model to datasets with a 200-nm, 300-nm or 400-nm z-step size. We performed training with 100,000 iterations and saved intermediate check-points every 5,000 iterations. For selecting a model for the different z-step sizes, we applied models from various check-points and took the best-performing check-point when evaluating F1 on the respective test datasets. For the 200-nm z-step size, we took the 70,000 and 50,000 iteration check-points for bassoon and SHANK2 prediction, respectively, and we applied the same model to the 400-nm z-step size. For the 300-nm z-step size, we used the 20,000 iterations check-point for both bassoon and SHANK2 prediction.
During the validation phase for the 300-nm z-step size prediction, training data comprised 5 of the 6 tiles (total volume of 14,207 µm3, with around 13,650 excitatory synapses), and one dataset was used for testing. For final training, all six datasets were included. During training, patches of training data (normalized between 0 and 1 with gp.normalize) were randomly sampled from the training datasets with equal probability and used in training if at least 0.1% of voxels were included in the binary mask generated during preprocessing of molecular signals (that is, 1,048.6 voxels of the 64 × 128 × 128-voxel patches). Training data were augmented by mirroring and transposing in the xy direction on structural and molecular channels. In addition, we applied intensity augmentation on the structural channel with (0.9, 1.1, −0.1, 0.1) for the two scaling and the two intensity shifting parameters, respectively, and enabled range clipping.
Prediction
Prediction was performed on ISTA’s high-performance computing cluster using SLURM for task allocation. We requested 8 CPUs, graphics acceleration by 1 GPU (NVIDIA GeForce RTX 2080 Ti or similar) and 100 GB of RAM. Prediction was performed in a piece-wise, ‘scanning’ mode, again with a patch size of 64 × 128 × 128 voxels after converting LICONN structural data to zarr format while applying the same normalization procedure as for the training data.
To convert predicted molecule locations to point annotations, we first rescaled intensity using the skimage.exposure.rescale_intensity, taking p1 and p99.95 as limits, with p1 referring to the first percentile and p99.95 to the 99.95th percentile. We then applied 3D Gaussian filtering with σ = 5 for bassoon and SHANK2. We applied Otsu thresholding to derive a binary mask and skimage.measure.label to extract instance segmentations. We erased volume segments smaller than 60 voxels and those that covered fewer than 13 imaging planes. We applied the skimage.measure.regionprops function to determine the weighted centroid for each segment on the basis of the intensity of the underlying LICONN structural channel, and placed a point annotation at that location.
Association of pre- and postsynaptic sites and validation of deep-learning-based synapse prediction were performed with the same pipeline as was used for immunolabelling-based synapse detection. The automated post-processing step for synapse detection was identical to the corresponding step in the immunolabelling-based synapse-detection pipeline and was applied to the test datasets (Fig. 4l–n and Supplementary Fig. 19).
Visualization
Neurite and synaptic segmentations, skeletons and imaging volumes were visualized in 3D with Blender versions 2.92, 3.2 or 3.51 (https://www.blender.org/). Video files were also generated with Blender. For visualizing neurites on an overview scale (Figs. 1a, 2c,d), precomputed meshes from Neuroglancer were used. Detailed views of 3D renderings (Fig. 1a zoomed dendrite, Figs. 1e,f, 3c) were based on meshes generated with the marching cubes implementation in scikit-image. For 3D rendering of molecular signals in the context of neuronal segmentations, immunolabelling channels and predicted molecule distributions were intensity scaled according to visual inspection, background was removed with σ ≈ 0.5 and σ ≈ 5 to preserve shapes and then Otsu thresholding was applied. To associate these signals with a particular neuronal segment, the segment was expanded with a ball of radius 6 and any molecular signals with at least 1 voxel overlap were retained. Meshes were again generated with the marching cubes algorithm.
Associating synapse detections with FFN-derived neuronal segments
To map detected synapses onto selected neuronal segments, we first expanded the segment with a ball of radius of 5 or 6 voxels. To map postsynaptic sites onto dendrites, we selected those detections whose centre coordinates were located within the expanded neuronal segment. We recalled the corresponding presynaptic partners using the matching matrix Mij to generate the list of full synapses associated with that segment. For axons, we first identified the pre-synapses located within the expanded segment, and then retrieved the corresponding post-synapses from the matching matrix. Owing to the large size of the imaging volume, computations were performed on manually defined patches, and results were combined for visualization and further analysis.
Automated analysis of connectivity based on deep-learning-based synapse prediction and neurite segmentation
To analyse connectivity in Fig. 4p, we first performed deep-learning-based synapse prediction on the imaging volume. For this, we predicted bassoon and SHANK2 molecule locations in smaller overlapping patches of the volume, and combined them into the final full-size prediction volume. Next, we converted the predicted molecule locations into synaptic point annotations using the algorithm described above. Because of the large size of the volume, pre-synapse and post-synapse annotations were computed on manually defined non-overlapping patches. Association of pre- and post-synapses—that is, computation of the matching matrix Mij—was performed on the entire volume to avoid errors in full synapse assembly owing to separation by a patch boundary. We then associated pre- and postsynaptic annotations with the neuronal segmentation.
For each pre-synapse (or post-synapse) location, we identified the underlying segment ID from the FFN-based neurite segmentation. If no segment was present at the coordinates of a specific pre- or postsynaptic detection, we performed an automated search in a single plane for the nearest non-zero segment in a 20 × 20-voxel neighbourhood. If (i) the squared distance between the respective pre-synapse (post-synapse) and the nearest non-zero segment was less than 30 voxel2 and (ii) the segment was classified as an axon (dendrite) by the neurite classifier described above, the respective pre-synapse (post-synapse) was mapped to the segment.
Finally, we combined the information from the matching matrix Mij and the synapse–segment associations obtained in the previous step to compute a matrix of connectivity between neuronal segments. For this, we iterated through the matching matrix Mij and added a connection between the segment associated with post-synapse i and the segment associated with pre-synapse j.
Statistics and reproducibility
GraphPad Prism v.10.1.2 was used for statistical analysis and plotting. Bootstrap analysis in Supplementary Fig. 21 was performed as described previously68, based on code available at https://gitlab.mpcdf.mpg.de/connectomics/human_primate. The violin plot in Fig. 2g was generated with seaborn.violin.plot (https://seaborn.pydata.org/generated/seaborn.violinplot.html). This is a proof-of-concept study focusing on the development of new technology and its applicability. Accordingly, no prior determination of statistical sample size was performed. Once the technology was established and the data quality was to our satisfaction, experiments were performed in multiple replicates to ensure and demonstrate reproducibility. For the comparison of primary cilia length in wild-type versus Hnrnpu+/− mice (Fig. 5g), we chose n = 3 biological replicates in each group, recorded across a total of n = 7 technical replicates to demonstrate that a biologically meaningful number of replicates can be analysed with LICONN. No randomization between experimental groups was performed. The comparison of cilia length between wild-type and mutant mice (Fig. 5g) was performed with an unpaired two-tailed t-test (P = 0.64; not significant). Measurements of cellular parameters (for example, distance measurements within synapses, periodicity and connectivity) were performed to show that quantitative information that is consistent with previous measurements can be extracted from LICONN data. Researchers were aware of the source of the data and the purpose of the study. Quantification of segmentation accuracy and synapse-detection fidelity were performed to evaluate how well computational algorithms could reproduce human ground-truth annotations. In these cases, blinding was not possible or appropriate. For determining the accuracy of human tracings relative to independent ground-truth information from sparse eGFP expression, blinding to that independent ground-truth information was performed. Human tracers were blinded to the eGFP channel for the analysis in Fig. 1e,f and Supplementary Figs. 13, 14. Blinding was implemented through the permissions settings in webKnossos.
Replication
To confirm the reproducibility of the technology, we performed a series of technical replicates, which were typically recorded across several biological specimens, as indicated below. For analyses, only datasets of high labelling and imaging quality were pursued. Typically, during the development phase, a number of experiments with some variation of parameters were performed. In all images, representative data from single experiments are shown. For some of the experiments that we performed routinely, the stated numbers give lower bounds and we did not count further replicates beyond n = 10. Fig. 1a,b: imaging data representative of acquisition of expansion and multi-tile imaging in n = 10 replicates across n = 3 mice. Fig. 1b represents subregion of dataset from Fig. 1a. Fig. 1c: 192 synapses analysed with multiple line profiles per synapse, in n = 3 technical replicates across n = 2 mice. Fig. 1d: 32 distance measurements across n = 2 mice. Fig. 1e and Supplementary Figs. 13, 14: 37 axon stretches analysed in 12 datasets from n = 3 technical replicates across n = 3 mice. Fig. 1f, Supplementary Fig. 15: 5 datasets from n = 3 technical replicates across n = 2 mice. Fig. 1g and Supplementary Fig. 6: single-tile LICONN measurements were performed with a large number of technical and biological replicates (n ≫ 20). Manual neurite annotation was performed in n = 2 datasets across n = 2 mice. Fig. 2a–i and Supplementary Fig. 16: multi-tile LICONN measurements were performed in n = 10 replicates. FFN-based segmentation was applied to n = 6 datasets. Comprehensive proofreading of all neuronal structures (Fig. 2a–g) and quantification of segmentation results using manually generated reference skeletons (Fig. 2h and Supplementary Figs. 16, 17) were performed in n = 1 dataset. Fig. 3a: immunolabelling for bassoon and PSD95 was performed in n = 3 technical replicates across n = 2 mice. Fig. 3b: analysis of 106 synapses in n = 3 technical replicates across n = 2 mice. Fig. 3c: visualization from the dataset in Fig. 3a. Bassoon and SHANK2 labelling was performed in n = 5 replicates across 3 mice. Fig. 3d: synaptic immunolabellings were replicated n = 2 times, with further immunolabellings for synaptic targets in various contexts. Fig. 3e: 185 synapses analysed in 3 imaging volumes from n = 1 technical replicate. Fig. 3f–h and Supplementary Fig. 19: immunolabelling-based, automated synapse-detection accuracy was evaluated against manually generated ground truth in n = 3 technical replicates recorded across n = 2 mice. Fig. 3f represents a schematic illustration of synapse detection (synapse annotations placed manually, n = 1). Fig. 3i: visualization of synapse detections was performed for n = 1 dendrite. Fig. 3j,k: immunolabelling for inhibitory synapses was performed in n = 5 technical replicates. Fig. 4a–k and Supplementary Figs. 20, 21: manual connectivity analysis was performed across 2 imaging volumes (total 154,560 µm3) in n = 1 technical replicate (n = 1 mouse). Fig. 4b–f: quantification of synaptic contacts of spiny dendrites was performed on n = 11 dendrites. Fig. 4h,i: output analysis and spine tracing was performed for n = 19 axons. Fig. 4k: output was analysed for n = 8 AIS-seeded axons. Fig. 4l–m and Supplementary Fig. 19: accuracy of deep-learning-based synapse prediction was evaluated against manually generated ground-truth annotations in n = 3 technical replicates recorded across n = 2 mice. Fig. 4n: mapping of synaptic inputs and outputs was performed in n = 1 cell and represents further analysis of the dataset in Fig. 1a. Fig. 4o: analysis of data in Fig. 2a (n = 1 dataset). Fig. 4p: visualization of excitatory and inhibitory inputs was done for n = 1 dendrite. Fig. 5a: immunolabellings were replicated n = 2 times in n = 2 mice. Fig. 5b: immunolabellings were replicated n = 3 times. Fig. 5c: imaging data representative of n = 10 replicates across n = 3 mice. Fig. 5e: immunolabellings were replicated n = 2 times. Fig. 5f: analysis of 78 cells from dataset in Fig. 1a. Fig. 5g: Hnrnpu+/−-haploinsufficient mice: 80 cells from n = 7 technical replicates across n = 3 mice; wild type: 78 cells from n = 7 technical replicates across n = 3 mice. Fig. 5h and Extended Data Fig. 9: representative of n = 2 replicates in n = 2 mice. Fig. 5i: Cilia diameter: 52 cross-sections; microtubule doublet distance: 54 distance measurements, analysed across n = 2 technical replicates in n = 1 mouse (5 imaging volumes). Fig. 5j and Extended Data Fig. 5: immunolabellings were reproduced in n = 2 replicates. Fig. 5k: representative of n = 3 replicates. Fig. 5l: representative of immunolabelling in n = 3 replicates. Fig. 5m: gap-junction and inhibitory-synapse density evaluated across n = 4 imaging volumes in cortex and hippocampus from n = 1 mouse. Extended Data Fig. 2: high-resolution large-scale tiling in hippocampus was performed in n = 1 replicate in n = 1 mouse. Extended Data Fig. 3: immunolabelling for RIM1/2 and VGLUT1 is representative of n = 2 technical replicates and bassoon and PSD95 immunolabelling of n = 3 replicates across n = 2 mice. Extended Data Fig. 5: the displayed combinations of immunolabellings for MBP and Cnx-43 were performed in n = 1 technical replicate, RIM1/2 and VGLUT1 in n = 2 technical replicates, GFAP and Cnx-43 in n = 2 technical replicates and vimentin and PMP70 in n = 1 replicate. Extended Data Fig. 6: LICONN imaging in different brain regions was performed in n = 1 mouse. Extended Data Fig. 7: overview imaging in CA3 stratum lucidum was performed in n = 2 technical replicates, single tile measurements were performed in multiple technical replicates. Extended Data Fig. 8: imaging of the various layers was performed in n = 1 mouse. Extended Data Fig. 9: data representative of replicates in n = 2 mice. Extended Data Fig. 10a: imaging data representative of LICONN in 300-µm-thick slices in n = 3 mice. Extended Data Fig. 10c–f: post-expansion hydrogel sectioning was performed in n = 3 technical replicates. Extended Data Fig. 10h: block-face imaging and sectioning over 12 rounds was performed in n = 1 specimen. Supplementary Fig. 1: data points correspond to individual technical replicates. Supplementary Fig. 1a: n = 1 replicate for 0.25%, 0.15%, 0.005%, n = 2 replicates otherwise. Supplementary Fig. 1b: different hydrogel recipes were tested in the following technical replicates: replicates 1–4 and 15: n = 2; replicates 5, 9, 11 and 13: n = 3; replicates 6–8, 10, 12 and 14: n = 4. Supplementary Fig. 1c: experiments were performed in the following technical replicates: 0.05% APS and TEMED: n = 3 replicates. 0.15% APS and TEMED: replicates 7, 8, 10, 11 and 14: n = 3 replicates; replicate 13: n = 2 replicates. Supplementary Fig. 1d: n = 2 technical replicates. Supplementary Fig. 1e: technical replicates: first hydrogel: replicates 11, 7, 14, 8 and 10: n = 3; replicate 13: n = 2. Final hydrogel: n = 4. Supplementary Fig. 1f: a single hydrogel was subdivided after the first gelation step. Expansion factors for the second expandable hydrogel were determined in the following technical replicates: replicate 11: n = 3; replicates 13, 7, 14 and 1: n = 4. Supplementary Fig. 1g: n = 4 technical replicates. Supplementary Fig. 2: data representative of n = 3 technical replicates. Supplementary Fig. 3: each condition was technically replicated n = 2 times. Hydrogels 14 and 15 were replicated n = 5 times. Supplementary Fig. 4a–c: individual data points represent individual mice. Number of mice analysed: Supplementary Fig. 4a–c: 20% AA: n = 3; 0% AA, 5% AA, 10% AA, 12.5% AA: n = 4; 15% AA: n = 6. Supplementary Fig. 4f: 0% AA: n = 15 cells across n = 1 technical replicate; 5% AA: n = 32 cells across n = 3 technical replicates; 10% AA: n = 98 cells across n = 3 technical replicates; 12.5% AA: n = 49 cells across n = 2 technical replicates; 15% AA: n = 74 cells across n = 3 technical replicates; 20% AA: n = 20 cells across n = 2 technical replicates; 25% AA: n = 20 cells across n = 2 technical replicates; 30% AA: n = 17 cells across n = 2 technical replicates. Supplementary Fig. 5: data representative of n = 3 technical replicates. Supplementary Fig. 6: manual segmentation in datasets with NAS anchoring was performed in n = 1 replicate. Supplementary Fig. 7: data representative of n = 3 technical replicates. Supplementary Fig. 8: data representative of n = 2 technical replicates. Supplementary Fig. 9: data representative of n = 3 technical replicates for each condition. Supplementary Fig. 10a: Measurement error represents mean ± s.d. at given measurement length from sampling 2 × 105 pairs of points in pre- and post-expansion images (see Methods) from the specific single measurement (n = 1 imaging volume) shown in the figure panel. Data representative of n = 4 technical replicates from n = 3 mice. Supplementary Fig. 10b,c: RMS measurement error over different measurement lengths for the first and second expansion steps and for the overall LICONN procedure were evaluated as mean ± s.d. across n = 4 technical replicates (using a total of 14 imaging volumes), recorded in cortex across n = 3 mice. Supplementary Fig. 10d: 21 imaging volumes analysed across n = 6 technical replicates from n = 3 mice. Supplementary Fig. 11: overview image in region corresponding to dataset in Fig. 1a (n = 1 dataset). Supplementary Fig. 12a,b: data representative of application of CLAHE in n = 10 technical replicates. Illustration of visual appearance of CLAHE-processed data at different slopes (Supplementary Fig. 12b) was performed in n = 1 dataset. Supplementary Fig. 18: data representative of n = 10 technical replicates. Visual illustration of resolution improvement by iterative expansion as shown in the figure was performed in n = 1 specimen. Supplementary Figs. 20, 21: manual tracings were performed across 2 imaging volumes from n = 1 specimen (n = 1 technical replicate) imaged in the high-resolution overview scan. Supplementary Fig. 22: tracing with deep-learning prediction of synapses was performed in n = 1 technical replicate. Supplementary Figs. 23, 24: immunolabellings were technically replicated n = 2 times. Supplementary Fig. 25: high-resolution overview imaging of the region analysed in Fig. 5g was performed in n = 1 specimen.
Creation of figures
Figures were created in Adobe Illustrator v.27.5 and v.27.7. The schematic in Fig. 5d and brain cross-section schematics in Supplementary Fig. 4 were created in BioRender (2025, https://BioRender.com/j05y134 and https://BioRender.com/x93f652).
Materials availability
Hnrnpu+/− mice are available from the authors upon request.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.