Mice
Young adult male and female mice were used between 2 and 4 months of age at the time of experimental procedures. C57BL/6 J mice (JAX: 000664) were used for experiments requiring a wild-type background. For RNA-seq of astrocyte-specific ribosome-associated mRNA mice expressing RiboTag43 (JAX: 029977) were crossed to the well-characterized, astrocyte-specific Cre-driver line, mGfap-cre 73.1237 to generate mGfap-cre-RiboTag mice. mGfap-cre-RiboTag mice were crossed to Stat3-loxP mice10 to generate mGfap-cre-Ribotag-Stat3-loxP mice (Stat3-cKO). Astrocyte-conditional Ccn1-knockout mice were obtained by crossing the well-characterized, astrocyte-specific Cre-driver line Aldh1l1-CreERT2 (ref. 31) JAX: 031008 to the Ccn1-LoxP line32 (a gift from K. Lyons) to generate Ccn1-cKO mice. Aldh1l1-CreERT2 mice were crossed to the floxed-STOP-tdT (Ai9) reporter line to generate Aldh1l1-CreERT2::floxed-STOP-tdT mice. Cre recombinase expression was activated in young adult mice (6–8 weeks old) by administering tamoxifen (Sigma, T5648-1G, 20 mg ml−1 in corn oil) by subcutaneous injection (100 mg kg−1, once a day) for 5 days followed by clearance for 3 weeks so that no residual tamoxifen remained at the time of experiment initiation. All mice were housed in a facility with a 12 h:12 h light:dark cycle and controlled temperature and humidity, and were allowed free access to food and water. Experiments were conducted according to protocols approved by the Institutional Animal Care and Use Committee at Cedars-Sinai medical centre.
Surgical procedures
All surgeries were performed on male and female young adult mice (8–12 weeks old) under general anaesthesia with isoflurane in oxygen-enriched air using an operating microscope (Zeiss), and rodent stereotaxic apparatus (David Kopf).
Spinal cord injury
Laminectomy of a single vertebra was performed at spinal cord level T12. Incomplete iSCI by unilateral T12 hemisection was performed on the left side of the spinal cord using a microknife (Fine Science Tools). To be included in the study, mice exhibited complete unilateral hindlimb paralysis for the first three days following surgery. A T12 crush SCI was made using no. 5 Dumont forceps (Fine Science Tools) with a 0.4 mm spacer and with a tip width of 0.5 mm. T12 crush mice exhibited paralysis in both hind limbs. In each case, mice received the opiate analgesic buprenorphine subcutaneously before surgery and every 12 h for 48 h after injury. Mice were evaluated thereafter blind to genotype and experimental condition. Daily bladder expression was performed for the duration of the study or until voluntary voiding returned.
Injections of lysolecithin or myelin into the spinal cord
Five-hundred nanolitres of 1% lysolecithin or 1 mg ml−1 CFSE-myelin in PBS was delivered by intervertebral microinjection to the lateral spinal cord white matter at spinal cord level T12 (coordinates: 200 μm medial–lateral, 300 μm dorsal–ventral). Injections were carried out at 150 nl min−1 using finely bevelled glass micropipettes connected via high-pressure tubing (Kopf) to 10 μl gastight syringes under the control of microinfusion pumps (Harvard Apparatus). Needles were left in place for 6 min prior to being slowly retracted. An equal volume of PBS was injected into the contralateral white matter as vehicle control. Mice were euthanized at 3 days post myelin injection and at 3, 10 and 25 days post-lysolethicin.
Sciatic nerve injury
A small incision was made on the left hindlimb and the two heads of the bicep femoris muscle were gently separated to reveal the sciatic nerve. The sciatic nerve was released from the muscle and elevated using forceps. The isolated nerve was then clamped with haemostats for 10 s and then replaced under the muscle. Mice were euthanized seven days following sciatic nerve crush.
Saporin injection
Conjugated saponins were used to degenerate myelinated and unmyelinated fibres as previously described44. In brief, mice were anaesthetised and 8 μg (10μl of 0.8 μg μl−1 in PBS) of saporin (non-conjugated control), IB4-conjugated saporin (targets unmyelinated fibres) or CTB-conjugated saporin (targets myelinated fibres) was injected subcutaneously into the plantar surface of the left hindpaw foot pad using a 30G insulin syringe. Injections of IB4–saporin and CTB–saporin were considered successful if there was local swelling in the treated hindpaw for 24–48 h following injection. Mice were euthanized at 14 and 28 days after injection.
EAE induction and assessment
Active EAE was induced as described45 with modifications. Nine-week-old C57BL/6 mice were immunized subcutaneously in both hind flanks with 100 μg of myelin oligodendrocyte glycoprotein peptide (MOG35–55) emulsified in Complete Freund’s adjuvant containing 200 μg of killed mycobacterium tuberculosis H37Ra (Hooke labs) and injected intraperitoneally on days 0 and 2 with 110 ng pertussis toxin. Assessment of EAE was as follows: 0, no disease; 1, decreased tail tone; 2, hind limb weakness; 2.5, partial hindlimb paralysis; 3, complete hind limb paralysis; 4, front and hind limb paralysis; and 5, moribund state. Mice were collected at different stages of disease on the basis of the following pre-defined criteria: onset, partial or completely limp tail (score 0.5–1) at day 10 ± 2 days; peak, near or complete paralysis of hindlimbs with or without forelimb weakness (score 2.5–3.5) at day 14 ± 2 days; chronic, mice that reached a score of at least 2.5 (limp tail and incomplete paralysis of hindlimbs) no later than day 16 and collected at day 56.
Myelin purification and conjugation
Myelin was purified from adult C57BL/6 mice brains by sequential ultracentrifugation on discontinuous sucrose gradient and hypo-osmotic shock as previously described25. Brains were homogenized with a glass Dounce in 10 mM HEPES, 5 mM EDTA and 0.32 M sucrose. This was layered on 0.85 M sucrose in HEPES/EDTA buffer and centrifuged in a SW41 Ti rotor at 24,600 rpm for 30 min with acceleration and deceleration set to 1. The crude myelin fraction was removed from interface, resuspended in ice-cold distilled water, and centrifuged at 9,500 rpm for 15 min. This step was repeated two more times. The pellet was then dissolved in 0.3 M sucrose in HEPES/EDTA buffer and placed on top of 0.85 M sucrose in HEPES/EDTA. All centrifugation/resuspension steps were then repeated. The final pure myelin pellet was resuspended in PBS, quantified using a BCA assay, and resuspended to 1 mg ml−1 and then conjugated to CFSE as previously described46. Myelin (1 mg ml−1) was incubated with 50 μM CFSE at 37 °C for 15 min and then washed with 100 mM glycine in PBS at 14,000 rpm for 15 min, washed twice with PBS at 14,000 rpm for 15 min each and pellets were then resuspended to 1 mg ml−1 in PBS.
Hindlimb locomotor evaluation
A modified Basso mouse scale (BMS) was developed to evaluate the gradual functional recovery of distinct hind limb muscle groups after iSCI, over time, in freely moving mice. We converted the original BMS protocol47 of 5 locomotor categories with a maximal score of 9 into 12 locomotor categories (ankle movement, toe movement, knee movement, weight support, paw placement, dorsal stepping, missing steps, paw position on lift-off, paw position on initial contact, coordination, trunk instability and tail tone) with a maximal score of 37. Analysis was performed at days −5, 0, 1, 2, 3, 7, 14, 28, 42, 56, 70 and 84 dpi.
Cold thermoception behavioural evaluation
Hindpaw sensitivity to cold stimuli was evaluated using the acetone test48. Spontaneous thermoceptive behaviours were monitored for 1 min after a drop of acetone (~25 μl) was applied to the plantar surface of left or right hindpaw with the aid of a 22G flexible gavage needle attached to a 1 ml syringe. The total duration of acetone-evoked behaviours (paw withdrawal, biting, licking or scratching) was measured from videos reviewed in slow motion. Analysis was performed at days −5, 7, 28 and 84 dpi.
Von Frey testing
Mechanical sensitivity was assessed using the von Frey filament test. Mice were placed in individual elevated chambers each measuring 3.75 × 3.75 × 5 inches with a mesh floor. Mice were acclimated to the testing chamber for 5 days before beginning any measurements and allowed to acclimate for 15 min prior to the start of testing on data recording days. A set of 20 nylon Semmes Weinstein monofilaments was used for testing. In brief, a fibre was gently pushed against the surface of the skin from below. Filaments of increasing stiffness (0.02–2.0 g) were applied perpendicularly to the plantar surface of the hind paw with sufficient force to cause slight bending and held for 2–3 s. A withdrawal response is categorized as an indicator of nociception and is defined as paw withdrawal, paw lifting, paw rotation, sniffing, licking, scratching, shaking or rapid movement, was recorded, and subsequent filament selection followed the up-down paradigm. If a withdrawal response was observed, the next lower force filament was applied; if no response occurred, the next higher force filament was tested. This process continued until six responses had been recorded in a series bracketing the threshold. Data were analysed using the up-down Reader algorithm to determine the 50% withdrawal threshold, which was calculated using Dixon’s formula.
Tissue processing, immunohistochemistry and mRNA in situ hybridization
Mice were euthanized by barbiturate overdose followed by cardiac perfusion with 4% paraformaldehyde. Spinal cords were removed, post-fixed for 4–8 h, and cryoprotected in buffered 30% sucrose. Spinal cords were blocked into 5 mm segments centred around the lesion epicentre, embedded in optimal cutting temperature (OTC) medium and stored at −80 °C until sectioning. Serial frozen sections of cervical (C8-T4), thoracic (T3-T12) and lumbar (T9–L3) segments (40 μm, transverse) were prepared using a cryostat microtome (Leica) and stored in antifreeze solution (glycerol, sucrose and TBS) at −20 °C until processed for evaluation by immunofluorescence and/or mRNA in situ hybridization as described9. Primary antibodies include: Rat-CD18 (1:100, Invitrogen), Rat-GFAP (1:1,000, Thermofisher), Rabbit-GFAP (1:1,000, Dako), Goat-IBA1 (1:1,000, Abcam), Rabbit-IBA1 (1:1,000, Wako), Rabbit-LPL (1:50, Abcam), Rabbit-PLIN2 (1:500, Progen), Goat-SOX9 (1:200, R&D system), Mouse- (1:3,000, Biolegend), Sheep-TREM2 (1:250, R&D systems), Rabbit-YAP1 (1:200, Protintech). Mouse primary antibodies were visualized using the M.O.M. (Mouse on Mouse) Immunodetection Kit (Vector Laboratories). Primary antibodies were selected on the basis of validation for fluorescence immunohistochemistry analysis in mouse tissue by the manufacturer, and/or by other investigators on the basis of peer-reviewed publications. Fluorescence secondary antibodies were conjugated to Alexa 488, Cy3 or Cy5 (all from Jackson Immunoresearch Laboratories). Nuclear staining was performed using DAPI (2 ng ml−1; Molecular Probes). Sections were cover-slipped using ProLong Glass mounting agent (ThermoFisher). When applicable, tissue sections were incubated in FluoroMyelin Green to label myelin and myelin debris (1:300) or the neutral lipid dye BODIPY to label lipid droplets (1:1,000) (ThermoScientific) prior to DAPI incubation.
Florescent in situ hybridization on fixed-frozen mouse spinal cord sections was performed using RNAscope probes and the Multiplex Fluorescent Detection Kit v.2 per manufacturer’s instructions (Advanced Cell Diagnostics). Mouse spinal cord sections were permeabilized with Protease IV. Probes used on mouse spinal cord tissue were as follows: Abca1 (522251), AldoC (429531-c2, 429531-c3), Arex (541871), Ak3 (454791), Boc (876211), Ccn1 (429001), Gfap (313211-c2, 313211-c3), Glipr2 (467171), Gpnmb (489511), Igf1 (443901-c2), Lair (509151), Prdm16 (584281) Scl1a3 (430781) and Thrsp (1090411). mRNAs of interest were labelled with the following fluorophores (Akoya): Opal 520 (FP1487001KT), Opal 570 (FP1488001KT), Opal 620 (FP1495001KT) and Opal 690 (FP1497001KT). Slides were then processed for immunohistochemistry or stained with DAPI before mounting. Human spinal cord tissue was permeabilized with target retrieval reagent and protease plus. Probes used in human tissue were as follows: CCN1 (4452081), GFAP (311801-C2) and SLC1AA3 (461081-C2). Sections were stained with DAPI and mounted with ProLong Glass or Vectashield mounting medium.
Reference to protein or gene names follow standardized guidelines for mouse and human as established by the Human Gene Nomenclature Committee (HGNC) and the Mouse Genome Informatics (MGI) database. Here, human and mouse proteins are referred to in all upper case, non-italicized font (for example, CCN1, IBA1, TREM2); genes and mRNA is referred in all upper case, italicized for human (for example, CCN1) and first letter capitalized and otherwise lower case, italicized for mouse (for example, Ccn1, Gpnmb, Igf1).
Imaging
Images of tissue sections used for quantitative analyses were collected using an Apotome epifluorescence microscope with structured illumination hardware and deconvolution software (Zeiss). For whole spinal cord Ccn1 and microglial analysis, we generated 10× tiles of the entire spinal cord at a single z-plane. Microglial quantification was imaged at 20× (Trem2, LPL) with a z-thickness of 1 μm or 40× (FluroMyelin, SMI32, PLIN2, BODIPY, Gpnmb and Abca1) with a z-thickness of 0.5 μm. Similarly, images of astrocytes with subtype markers and were imaged at 40× with a 0.5 μm z-stack. Representative images for illustrative purposes were imaged on a Leica SP7 Confocal microscope at 20× or 63×.
Image analysis
Imaris image analysis software (v.10) was used to generate 3D volumes of surfaces of IBA1+ microglial and a marker of interest (for example, FluroMyelin, SMI32, PLIN2, BODIPY, TREM2). Overlap between IBA1 and marker surfaces (≤0.5 μm distance) was used to determine the proportion of microglia that were marker-positive. Similarly, overlap of marker-positive surfaces that were within an IBA1 surface (≤0.5 μm: TREM2, PLIN2, BODIPY, FluroMyelin, SMI32) determined the volume of marker present within microglia. Measurements were normalized to the total volume to IBA1 microglia and measurements were restricted to the spinal cord dorsal white matter unless stated otherwise. For YAP1 analysis, 3D surfaces were generated for all DAPI+ nuclei, YAP1, Gfap/Slc1a3 mRNA and Ccn1 mRNA. Astrocyte nucelli were determined by setting the overlap volume of DAPI and Gfap/Slc1a3 to 15. Astrocytes nuclei expressing Ccn1 were those containing an overlap volume of Ccn1 greater than 0.16. Finally, the YAP1 expression within the Ccn1+ and Ccn1− astrocytes was the volume of YAP1 (<0 μm) within these Ccn1+ or Ccn1− astrocyte nuclei.
Spatiotemporal analysis of Ccn1+ astrocytes and microglial nodules
Regional quantification of Ccn1+ astrocytes and IBA1+ microglial nodules was performed on 10× image tiles of transverse spinal cord sections using the cell counter plugin (Fiji). Transverse sections were only evaluated if they appeared cytoarchitecturally intact with normal-appearing white and grey matter anatomy. Initially, 8 anatomical reference points were used to align transverse spinal cord images: central canal; top of the dorsal white matter; bottom of the dorsal white matter; left and right lateral white matter; top and bottom of the central grey matter; the left and right sides of the central grey matter; the top of the dorsal horn grey matter on left and right sides, and the bottom of the ventral horns on left and right sides. For injured samples, the side containing the majority of Ccn1+ astrocytes or microglial nodules was labelled as left (ipsilesional). Next, Ccn1+ astrocytes were quantified as Gfap/Slc1a3 containing nuclei that contained at least 3 Ccn1 mRNA puncta (RNAscope). Similarly, microglial nodules were quantified as closely associated clusters of microglia containing more than three microglial nuclei25. At least two sections were quantified per mouse. Ccn1+ astrocytes and microglia nodule counts from different tissue sections were aligned to a common coordinate system using a custom python script. First, all reference and cell coordinates were linearly shifted such that the central canal was set at (0,0). The average of each reference point across all sections per spinal region were used to define a template spinal section which was then used to perform non-rigid transformation (ThinPlateSplineShapeTransformer from the OpenCV2 library) of all cell coordinates. For visualization, Ccn1 astrocyte/microglia counts were spatially binned per section using a 2D histogram (bin area 19.35 μm2) and counts per bin were averaged per mouse and then per condition. The resulting cell count per bin was then plotted.
For statistical comparison of time-dependent differences in WDM nodule formation between wild-type and Ccn1-cKO mice, we employed a Conway–Maxwell–Poisson (COM–Poisson) generalized linear mixed model with a log link and fixed effects for group and time49. This model was selected to enable robust assessment of how nodule counts change over time in wild-type and Ccn1-cKO mice, and whether these changes differ by genotype. The COM–Poisson distribution accommodates both overdispersion and underdispersion in count data and accounts for the repeated-measures structure of the dataset, wherein multiple tissue sections were analysed per biological replicate at each post-injury time point. The model included fixed effects for group (Ccn1-cKO versus wild type), time (7, 28 and 90 dpi), and a random intercept for each mouse to account for within-subject clustering. The group-by-time interaction was formally tested using a likelihood ratio test comparing nested models with and without the interaction term. Post hoc pairwise comparisons of group (genotype) and time levels were conducted on the basis of model-derived estimated marginal means, with multiple testing adjustment using Tukey’s method. All hypothesis tests were two-sided with a significance level set at 5%. These statistical analyses were performed using R software (v.4.4.1). Mixed models were fitted using the glmmTMB package50, and marginal means were estimated using the emmeans package51.
Quantitative analysis of in situ mRNA hybridization
Quantification of RNAscope probe signal (mRNA) in astrocytes and microglia was carried as described52. In brief, thresholding of RNAscope probe signal was first carried out (Otsu method: Ccn1 and Gfap/Slc1a3; triangle method: Gpnmb and Abca1) and the area of pixels was then quantified within the soma of Gfap+/Slc1a3+ astrocytes, or IBA1+ microglia or microglial nodules, respectively. The area of Gfap/Slc1a3 and Ccn1 were analysed from the same astrocyte somas, whereas the area of Gpnmb, Abca1 and Sdc4 mRNA was then normalized to the size of the microglia or nodule.
Fresh spinal cord tissue collection for astrocyte RiboTag RNA-seq
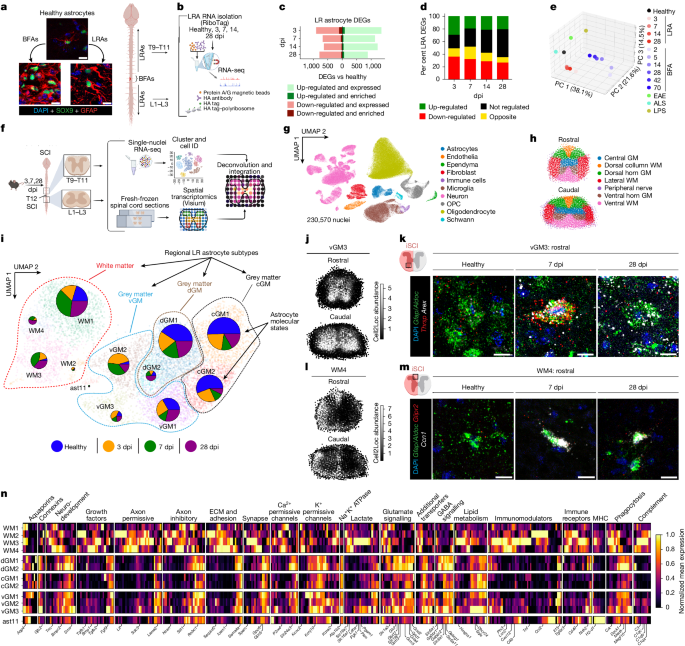
Spinal cord tissue was isolated for astrocyte RiboTag RNA-seq as described9. In brief, wild-type (mGfap-cre-RiboTag) and Stat3-cKO (mGfap-cre-Ribotag-Stat3-loxP) mice were perfused with ice-cold PBS with heparin and spinal cords were dissected out. Three millimetres of spinal cord rostral (T9–T11) and caudal (L1–L3) to the lesion epicentre were then rapidly removed, snap-frozen in dry ice and stored at −80 °C until processing for RiboTag RNA-seq. Spinal cords were collected at 3, 7, 14 and 28 dpi and anatomically equivalent regions of spinal cord were isolated from age- and genotype-matched healthy controls.
Astrocyte ribosome-associated mRNA isolation, RNA-seq and analysis
Astrocyte ribosome-associated mRNA was isolated using our previously established methods9. In brief, fresh frozen spinal cord tissue was homogenized and haemagglutinin (HA) immunoprecipitation was carried out to purify of astrocyte ribosome-associated mRNA. Astrocyte RNA integrity was analysed using the 2100 Bioanalyzer (Agilent) with the RNA Pico chip, with RNA integrity number (RIN) ≥ 8 for all samples. RNA concentration was determined using the RiboGreen RNA Assay kit (Life Technologies). cDNA was generated from 10 ng of RNA using the Universal plus mRNA-seq Kit (Nugen). The workflow consisted of poly(A) RNA selection, RNA fragmentation and double-stranded cDNA synthesis using a mixture of random and oligo(dT) priming, followed by end repair to generate blunt ends, adaptor ligation, strand selection and PCR amplification to produce the final library. Multiplexed sequencing was performed using the NovaSeq 6000 sequencer (Illumina) on a NovaSeq S2 flow cell to produce 50 bp paired-end reads. Data quality was assessed using Illumina SAV and demultiplexing was performed using Illumina Bcl2fastq2 v.2.17. Sequences were aligned to the mouse mm10 genome using STAR aligner (v.2.4.0j). Average percent of uniquely mapped reads was 79 (±8.7)%. Read counts were determined using HT-seq (v.0.6.0). At least 4, and in most cases 6 samples were evaluated per experimental condition. Genes not expressed in minimum of 10 samples (5 counts or more) or average fragments per kilobase per million mapped fragments (FPKM) below 0.75 were filtered out from further analysis. Differential expression analysis (DEA) was conducted using the Bioconductor EdgeR package (v.3.6). DEGs were determined using FDR at 5%. To identify co-regulated astrocyte-enriched genes across time after injury, a gene-gene correlation matrix was constructed using genes that were significantly enriched in astrocytes with a logFC >1 and FDR P ≤ 0.05 at any time point. Astrocyte-enriched gene expression was identified by comparing astrocyte HA immunoprecipitation-derived ribosome-associated mRNA to whole-tissue mRNA (HA immunoprecipitation input-derived mRNA). Astrocyte versus whole-tissue DEA identified 1249 astrocyte-enriched DEGs, which were used as input for a spearman correlation using log2FC changes values from iSCI versus healthy DEA and kmeans clustered into 11 gene modules. Genes in each module were used as input into gene ontology (GO) using Enrichr (GO_Biological Process_2018 database).
LRA gene expression data were compared to multiple other spinal cord astrocyte transcriptomics data sets from SCI and other non-traumatic CNS insults and disorders: BFAs17, ALS53, LPS9 and EAE54. A composite list of DEGs (log2 fold change versus healthy) across all datasets was compiled (9,558 DEGs) and used for all downstream analyses. Principal component analysis was performed as a descriptive visualization of global relationships across datasets, in conjunction with gene-level analyses of DEG overlaps, directionality, and functional groups. Together, these complementary approaches allowed us to assess both overall transcriptomic relationships and specific gene-level distinctions. For comparison between LRAs and BFAs, LRA 3 and 7 dpi were compared to BFA 2 and 5 dpi, respectively. DEG information was available for 14 and 28 dpi in both datasets. These data were used to tabulate the proportion of LRA DEGs that were either also significantly upregulated, downregulated, oppositely regulated, or not regulated in BFAs at each time point.
Nuclei isolation
iSCI mice were perfused with ice-cold PBS with heparin at 3, 7 or 28 dpi, spinal cords dissected out and 3 mm of spinal cord rostral (T9–T11) and caudal (L1–L3) to the lesion epicentre were then rapidly removed, snap-frozen in dry ice and stored at −80 °C. An anatomically equivalent region of spinal cord (T11–L1) was isolated from age- and genotype-matched healthy controls. Frozen tissue was homogenized in homogenization buffer (320 mM sucrose, 0.1 mM EDTA, 0.1% IGEPAL CA-630, 5 mM CaCl2, 3 mM magnesium acetate, 10 mM Tris, Roche Protector RNAse Inhibitor, Complete Roche Protease Inhibitor v.12, 0.016 mM PMSF, 0.166 mM β-mercaptoethanol; pH=7.8). Nuclei were isolated from the homogenate by iodixanol gradient and resuspended in 1% BSA solution before proceeding immediately to 10x snRNA-seq.
snRNA-seq
snRNA-seq was performed using 10x Chromium Next GEMSingle Cell 3 (v.3.1) per manufacturer’s instructions. Samples were loaded to capture 10,000 nuclei per sample. During library preparation, the initial cDNA amplification was run for 13 cycles, which was found to be optimal for 10,000 nuclei. Following library preparation, quantitative PCR was run to quantify library concentration and samples were pooled to equivalent concentrations. Initially, a shallow sequencing run of the pooled libraries at ~20% sequencing saturation, the results of which informed library re-pooling in order to normalize nuclei number within the libraries to obtain ~40,000 reads per cell. Sequencing was performed by NovaSeq (Illumina) at 2 × 150 base pair reads at 150 pM (average reads per sample: mean: 2.9 × 108 ± 1.1 × 108).
snRNA-seq data analysis
Output FASTQ files for each sample were aligned with CellRanger v.6.0.2 using the mm10-2020-A reference genome for each sample. Cells matching the following criteria were removed from further analysis: >5% mitochondrial counts, >25,000 counts or <500 counts. Genes expressed in fewer than 50 cells were removed from downstream analysis. Scrublet55 was used to remove predicted doublets from each sample. Individual sample data were then concatenated, normalized to 104 total counts per cell, log-transformed, and batch corrected using Harmony56. Quality control thresholding resulted in 230,620 cells from 35 samples for downstream analysis. Cell types were identified on the basis of putative marker genes12,57,58,59. DEG testing utilized sc.tl.rank_genes_groups with method=‘wilcoxon’ and corr_method= ‘benjamini-hochberg’ for all comparisons. Molecular markers of regionally restricted LRA subtypes (such as vGM3: Thrsp/Arex; WM4: Glipr2/Ccn1; see Fig. 1) were identified by screening our snRNA-seq data for genes that were (1) astrocyte-enriched relative to other cell types; and (2) significantly enriched for in a spatially restricted LRA subtypes. Nichenet30 was performed on astrocytes (‘sender’) and ligands were identified by filtering Nichenet candidates for astrocyte subcluster enrichment relative to all other cell types. The relevant receiver cell type was selected on the basis of NMF cell subtype enrichment. Genes enriched in receiver cell subtype were used as gene set of interest. All expressed genes in the receiver cell subtype were used as the background gene set.
Spatial transcriptomics
Mouse spinal cord spatial transcriptomics was performed by Visium (10x Genomics). iSCI mice were perfused with ice-cold PBS with heparin at 3, 7 or 28 dpi, spinal cords were dissected out and rostral and caudal blocks were rapidly embedded in OTC, snap-frozen on dry ice and stored at −80 °C until sectioning. Visium slides were pre-chilled in a cryostat (Leica) for 30 min at the time of sectioning. Two 10-μm sections were taken from lesion-remote rostral (T9–T11) and caudal (L1–L3) blocks, equivalent to samples analysed by snRNA-seq. Samples were processed using the Visium Spatial Gene Expression Reagent Kit (10X Genomics) per the manufacturer’s established protocol. cDNA libraries were pooled in a NovaSeq6000 SP v.1.0 flowcell and paired-end sequencing was performed on an Illumina NovaSeq6000 sequencer.
Spatial transcriptomics analysis
Spots overlaying tissue sections were manually annotated in the Loupe (10X Genomics) and processed by spaceranger-v.1.3.0 and aligned against the mm10 reference genome mm10-2020-A. Haematoxylin and eosin staining of transverse spinal cord sections was used to manually annotate lateral white, ventral white, dorsal white, central grey, dorsal horn and ventral horn. Additionally, gene expression of inflammation and gliosis-associated genes was used to distinguish lesion ipsilateral and contralateral sides. Quality control thresholding resulted in 14,566 spots across 16 biological replicates (n = 4 mice per group; 2 sections per rostral and caudal block). Data were normalized to 104 counts and log-transformed before running principal component analysis and UMAP projection. To accommodate for morphological variation, a non-rigid transformation was applied (ThinPlateSplineShapeTransformer from the OpenCV2 library) using manually placed neuroanatomical reference points, in a manner equivalent to aligned average density plot construction for Ccn1+ astrocyte and WDM nodules counts. Tissue alignment was validated by examination of known spatially restricted gene expression (for example, Mbp and Syp). Cell2location was used to spatially integrate snRNA-seq subclusters and spatial transcriptomics data60. The top 30 highest expressed genes from each dataset, mitochondrial genes, and genes expressed in <5% of cells or spots and with mean <1.12 were filtered out to generate snRNA-seq input. Cell2location was run with the following parameters: (batch_key= “Date library prep”, continuous_covariates = “total_counts”, categorical_covariates = “User”, N_cells_per_location=12, detection_alpha=200) and trained for 40,000 epochs. The cell2location matrix was used as input for NMF to identify spatially co-occurring cell types. NMF from sklearn was run with the following parameters: (n_components=8, alpha=0.9,max_iter=1000, shuffle=True, init = “nndsvda”,l1_ratio=0.9).
Microglial isolation (lipidomic and culture)
Mice were perfused with ice-cold PBS with heparin and the brain and spinal cord were freshly dissected following. For iSCI mice, 1 mm rostral and caudal to the lesion epicentre was removed and discarded, and the injured lateral side of the spinal cord (rostral and caudal to the lesion) was collected for microglial isolation. Dissected tissue was minced with a sterile razor blade and then dissociated using the Neural Tissue Dissociation Kit (P) (Miltenyi Biotech) and the GentelMACS dissociator with heaters per the manufacturer’s instructions. Following dissociation, samples were filtered through a 70-μm strainer, and myelin was depleted using Myelin removal beads II (Miltenyi Biotech) and the AutoMACS separator. Finally, microglia were isolated by incubating samples with CD11b microbeads (Milteyni) and isolating with the AutoMACS per the manufacturer’s instructions. Cell number was then determined before proceeding to downstream applications (lipidomics and culture).
Microglial bulk RNA-seq and analysis
A 50 μl spot drop containing 50,000 microglia cells was seeded into the desired wells of a 24-well plate. Cells were cultured for 7 days, with a full media change given on day 1 followed by half media change on day 4. On day 7, cells were either left untreated or treated with CCN1 (Peprotech: 120-25) at 50 ng ml−1 or vehicle (BSA) for 24 h. Each condition was carried out in triplicate for each experiment. Cells were then removed using a cell scraper, replicates pooled, and then processed for RNA isolation using RNeasy Plus Micro Kit (Qiagen, 74034). Microglia RNA samples were prepared for analysis by RNA-seq. RNA integrity was analysed using the 2100 Bioanalyzer (Agilent) with the RNA Pico chip, with RIN ≥ 9.6 for all samples. RNA concentration was determined using the RiboGreen RNA Assay. Multiplexed paired-end sequencing was performed using the NovaSeq X Plus sequencer (Illumina). Data quality was assessed using Illumina SAV and demultiplexing was performed using Illumina Bcl2fastq2. Sequences were aligned to the mouse mm39 genome using STAR aligner (v.2.7.11b). DEA was conducted using EdgeR with a FDA of 5%. DEGs were defined with a log2-transformed fold change threshold of 0.25 and adjusted P value < 0.05 (CCN1 stimulation versus vehicle).
Microglia lipid efflux and lipid droplet content evaluation
Primary microglia isolated from male and female mice 8–12 weeks of age were seeded at 1 × 105 cells per well in a flat-bottom 96-well plate coated with poly-L-lysine (0.01%, Sigma-Aldrich). Cells were grown in microglia media (10% fetal bovine serum, 1× penicillin-streptomycin, 10 ng ml−1 carrier-free (CF) recombinant mouse GM-CSF, 10 ng ml−1 CF recombinant mouse M-CSF, 10 ng ml−1 CF recombinant human TGFβ1, in DMEM/F-12 Ham) for 4 days at 37 °C, 5% CO2 with full media change after 24 h. Microglia cholesterol efflux was assessed using a Fluorometric Cholesterol Efflux Assay Kit (Abcam; ab196985) following the manufacturer’s instructions. In brief, on day 4 microglia were loaded with fluorescent cholesterol for one hour, then placed into equilibration media containing CCN1 (50 ng ml−1 in 0.1%BSA) or vehicle (0.1% BSA) for 16 h. Following incubation, cells were washed with phenol red-free DMEM/F-12 Ham and incubated with cholesterol acceptor solution (2% (2-Hydroxypropyl)-β-cyclodextrin) for 6 h. Plates were then centrifuged for 2 min at 1,000g and the cell supernatant was collected for fluorometric analysis of cholesterol content. Meanwhile, adherent cells were lysed and processed for fluorometric cholesterol content within the cell. Fluorometric measurements were performed using Varioskan LUX (ThermoFisher) (excitation/emission, 485/523 nm). Per cent cholesterol efflux was then calculated for each sample by dividing relative fluorescence unit (RFU) of the supernatant by the total cholesterol content (RFU of supernatant plus cell lysate). The effect of treatment was then calculated by subtracting the percent cholesterol efflux of the negative control (no cholesterol acceptor) from the percent cholesterol efflux of treatment (CCN1 or vehicle), followed by normalization with the percent cholesterol efflux of vehicle.
For microglial lipid storage analysis, primary microglia were seeded at 500,000 cells per well in 6-well plates and cultured for 7 days, with a full media change on day 1 and a half media change on day 4. On day 7, cells were treated with purified recombinant CCN1 (50 ng ml−1; Peprotech) or vehicle control (BSA) for 22 h. To assess the involvement of SDC4, cells were pre-treated with either SDC4 function-blocking antibodies (Rat Anti-Mouse SDC4 Clone KY/8.2, BD Pharmingen, 550350) or serotype-matched control antibodies (Rat IgG2a kappa Isotype Control, eBioscience, 14-4321-82) (25 µg ml−1) for 1 h. Media containing antibodies was then washed out and replaced with media containing CCN1. After 22 h, culture media was removed and cells were labelled with LipidTOX Deep Red Neutral Lipid stain (Thermo Scientific, H34477; 1:500 in PBS) for 30 min. Cells were washed with PBS-EDTA, gently dissociated by pipetting, and collected by centrifugation (300g, 10 min, 4 °C). Cell pellets were resuspended in 500 µl of flow cytometry buffer (PBS + 5% FBS), stained with DAPI, and analysed on a SONY ID7000 Spectrum Cell Analyzer at the Cedars-Sinai Medical Center Flow Cytometry Core. FlowJo software was used for downstream analysis. The gating strategy used to identify and quantify microglia populations is provided in Supplementary Data 6.
Sample preparation for lipid extraction
Lipid extraction from the frozen microglial cell pellets was done following Bligh and Dyer protocol with slight modifications61,62. In brief, the pellets were thawed at 4 °C for 10 min, after which, cold methanol, and HPLC-grade chloroform were added in a ratio of 2:1. The samples were vortexed for 10 s, resulting in a one-phase solution which was then incubated for 15 min at 4 °C. A biphasic solution was then obtained by adding ultrapure water and chloroform in a 1:1 ratio. Next, the samples were centrifuged at 16,000g for 10 min, giving rise to 3 phases in each tube. The bottom phase in the tube is the organic phase that contains lipids. Next, the solvents from the organic phase were evaporated using SpeedVac vacuum concentrator for 1 h resulting in dried lipid extracts.
Unbiased Lipidomics using MRM profiling
Dried lipid extracts from microglial cells were reconstituted in 200 µl of a 50:50 methanol:chloroform solution containing 10 mM ammonium formate. Prior to analysis, lipid extracts were further diluted in acetonitrile: methanol 70:30, with 10 mM ammonium formate. The quality control sample was the injection solvent containing 0.02 µg ml−1 of the quantitative mass spectrometry internal standard EquiSPLASH (Avanti Polar Lipids, 330731), which was monitored over time to ensure the instrument’s appropriate operation. All MRM profiling experiments were conducted on an Agilent 6495C triple quadrupole mass spectrometer outfitted with an Agilent 1290 Infinity II LC system and G7167B autosampler. A volume of 8 μl of diluted lipid extract was introduced into the Agilent Jet Stream (AJS) ion source of the mass spectrometer by flow injection for each MRM method. In brief, MRM methods were established for 10 lipid classes and spanned 1,497 individual lipid species62,63. Lipid classes of interest were phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylglycerol (PG), phosphatidylinositol (PI), phosphatidylserine (PS), acyl carnitine (CAR), cholesterol ester (CE), diacylglycerol (DG), triacylglycerol (TG), and sphingomyelin (SM).
Lipidomics data analysis
Statistical analysis of lipid MRM transitions for all sample comparisons was performed using the CLAW in-house MRM processing software64, followed by differential analysis with the edgeR software package65, as described in our previous study62,63. The ion count of each lipid was denoted by l for a given sample s. An intercept sample, representing the experimental blank (injection medium), was included to ensure that all comparisons are meaningful relative to the blank. The expected count for lipid l in sample s was written as μls. The design matrix entry \({X}_{s}^{T}\) encoded the group or condition for sample s, and βl represented the set of regression coefficients for lipid l. The edgeR package fits a generalized linear model to a log-linear formula for mean variance relationship as follows:
$$\log {\mu }_{{l}{s}}={X}_{s}^{T}{\beta }_{l}+\log {N}_{s}$$
This formula calculates the total ion intensity for each sample s, summing to Ns. This approach enables the determination of the coefficient of variation (CV) for the ion count of each lipid in a sample (yls). The dispersion (Φl) of each lipid and is calculated using the common dispersion method5. On the basis of these values, the log2 fold change (log2FC) between samples is calculated, and the corresponding P values are derived using the likelihood ratio test. P values less than 0.05 were considered significant. Microglia lipidomics data are available online at https://github.com/chopralab/Lesion-remote_astrocytes_govern_microglia-mediated_white_matter_repair.
$${\rm{C}}{{\rm{V}}}^{2}({y}_{{ls}})=1/{\mu }_{{ls}}+{\varPhi }_{l}$$
CCN1 co-immunoprecipitation
To identify candidate CCN1 receptors on microglia, a CCN1-directed bait-and-prey approach was used. Co-immunoprecipitation was performed using the Pierce Crosslink Magnetic IP/Co-IP Kit (Thermo Fisher Scientific, 88805) following the manufacturer’s protocol with minor modifications. In brief, 25 µl of protein A/G magnetic beads were resuspended, washed on a magnetic stand, and incubated with 5 µg of anti-DDK for 2 h at 4 °C for conjugation. Beads were incubated with 5 µg IgG isotype control antibodies for negative control. Bead–antibody conjugates were then washed twice with 500 µl of wash buffer.
HEK-293T cells expressing Mouse CCN1 with a DDK–MYC molecular tag (Origene MR221828) were lysed in buffer supplemented with protease inhibitors, followed by centrifugation at 12,000g for 10 min to remove debris. The cleared lysate was incubated with bead–antibody conjugates overnight at 4 °C on a rotator to allow CCN1 binding. The following day, the beads were washed three times with 500 µl of wash buffer before incubation with microglial lysates. Next, primary microglia were cultured for 7 days then lysed by sonication in sample buffer (50 mM Tris, 150 mM NaCl, 1% NP-40 (v/v), 0.5% CHAPS (w/v), protease and phosphatase inhibitors, pH 7.4). The whole-cell lysate was then incubated with bead–antibody–CCN1 complexes overnight at 4 °C on a rotator. Bead–antibody–CCN1 complexes were then washed three times with wash buffer and sent to the Cedars-Sinai Proteomics and Metabolomics Core for on-bead digestion and proteomic analysis by liquid chromatography–mass spectrometry (LC–MS/MS).
Sample preparation for proteomics
Samples were lysed in 6 M urea, 1 M ammonium bicarbonate, 5% SDS lysis buffer and sonicated for 10 min at 70% power using a QSonica Q800 sonicator. Samples were cleared by centrifugation at 20,000g for 10 min and protein concentration measured by BCA. Samples were digested by an automated SP3 protocol adapted to a Beckman i7 workstation. Bead aliquoting, reduction, alkylation, digestion, and elution were all performed on-deck with a 96-well plate format. In brief, 50 μg of protein in 40 μl of the previously mentioned lysis buffer was reduced with the addition of 10 μl of 200 mM dithiothreitol and incubated 30 min at 37 °C with shaking at 300 rpm, then alkylated with 10 μl of 400 mM iodoacetamide at room temperature for 15 min in the dark. The volume was brought to 70 μl with Tris-HCl pH 8, then 5 μl of bead suspension (10:1 mass ratio of beads to protein), 1:1 mixture of hydrophilic/hydrophobic beads (Cytiva) was aliquoted into the samples using the span-8 pipetting head with constant agitation of the bead reservoir between transfers. Samples were brought to 50% acetonitrile (ACN) and incubated for 18 min, and then the solvent was removed on-magnet, and samples were rinsed with 2× 80% ethanol then 2× ACN with 200 μl volumes each. After the solvent was completely removed, the samples were resuspended in 50 mM Tris-HCl pH 8 and 10 mM CaCl2 with trypsin at a 1:20 ratio. Samples were bath sonicated for 5 min then incubated 18 h at 37 °C and 1,200 rpm overnight. After digestion, the samples were then removed from beads and brought to 0.1% formic acid and 2% DMSO for injection into the instrument.
LC–MS/MS analysis
Approximately 500 ng of peptides from digested samples were analysed on a Thermo Orbitrap Astral coupled to a NeoVanquish LC. To assess carryover, a blank injection was included after every three injections. Samples were separated using 24 min gradient. The compositions for solvent A and B were 0.1% formic acid in water and 80% ACN with 0.1% formic acid, respectively. The gradient used was as follows: 1.2 μl min−1 flow, 0 min 4% B, 2 min 9% B, 13 min 25% B, 17 min 35% B. NeoVanquish LC was operated in direct injection mode, using a 150 μm internal diameter × 15 cm, 1.5 μm PepSep C18 (Bruker) column coupled to a nano source (Thermo EasySpray) on the Orbitrap Astral MS platform (ThermoFisher). All sample runs were acquired in data independent acquisition (DIA) mode from 380 to 980 Da with 240k Orbitrap resolution and 5 ms maximum injection time for MS1. All DIA scans were set to a 7 ms maximum injection time with varying window schemes between 2 and 5 Th depending on gradient length.
Proteomic data analysis
Mass spectrometry raw data files were searched against UniProt mouse reviewed protein sequence entries (accessed April 2023) using DIA-NN (v.1.8.1) (PMID: 31768060) in library-free mode with default parameters. On the basis of recent comparisons with library-based approaches, DIA-NN in library-free mode has been found to produce results that are comparable or better than those of experimental library-based searches while being freely available and was hence chosen for the analysis of all data. (PMID: 36609502). The output protein group matrix from DIA-NN was used to perform downstream analysis using MetaboAnalyst 6.0 (PMID: 38587201). Pairwise comparisons between CCN1 co-immunoprecipitation and negative control antibody co-immunoprecipitation were performed using uncorrected two-sided t-tests, with a significance threshold of −log10(P) > 1.3 (P < 0.05). No multiple hypothesis correction was applied given the exploratory nature of the analysis.
Adult mouse cortical astrocyte culture and conditioned media
Adult mice were perfused with ice-cold PBS with heparin and mouse brains freshly dissected. Cortices were dissociated using Neural Tissue Dissociation Kit (P) (Miltenyi Biotech) and the GentleMACS dissociator with heaters per the manufacturer’s instructions. Following dissociation, samples were filtered through a 70-μm strainer, and myelin was depleted using 120 μl of Myelin removal beads II (Miltenyi Biotech) in 1,000 μl of MACS Buffer (0.5% BSA, 2 mM EDTA in PBS) using LS columns (modified from PMID: 26919701). The myelin-depleted sample was then treated with Debris Removal Solution (Miltenyi Biotech) per the manufacturer’s instructions to remove any further cellular debris. Finally, astrocytes were isolated using ACSA-2 beads with LS columns per the manufacturer’s instructions (Miltenyi Biotech). Astrocytes were resuspended in AstroMACS media (Miltenyi Biotech) and plated at 100,000 cells per well on laminin and poly-l-lysine coated coverslips and incubated at 37 °C with 5% CO2. A half media change was performed every two days per the manufacturer instructions. On day 8 astrocyte conditioned media was collected, centrifuged (300g, 10 min, 4 °C) and the supernatant concentrated using 10 kDa Amcon Ultra Spin Columns (4,000g, 30 min, 4 °C). Concentrated astrocyte conditioned media samples were stored at −80 °C until analysis.
Western blot
Spinal cords were isolated from healthy and 3 dpi adult mice (n = 4, male and female) to evaluate CCN1 protein levels. Lesion-remote spinal cord tissue rostral and caudal to the SCI lesion and equivalent regions from the healthy core were homogenized by Dounce homogenizer using ice-cold RIPA lysis buffer (Thermo Scientific 89900) with cOmplete, Mini, EDTA-free Protease Inhibitor Cocktail (Roche, 04693159001). Homogenates were centrifuged at 17,000g for 30 min at 4 °C, and the resulting supernatant was collected. Protein concentration was estimated by Pierce BCA Protein Assay Kits and Reagents, (Thermo Scientific 23225). Total protein from astrocyte conditioned media was precipitated with acetone (−20 °C for 2 h) and centrifuged. The precipitated proteins were then centrifuged at 13,000 rpm for 10 min, and subsequently dissolved in RIPA buffer. Finally, samples were mixed with Laemmli sample buffer, heated, and resolved via SDS–PAGE for subsequent western blotting.
In each case, equal amounts of protein were resolved by 10% polyacrylamide gel electrophoresis (Tgx FastCast Acrylamide Kit, 10%, 1610173, Bio-Rad laboratories) with Precision Plus Protein Blue-Stained Protein Standards, 10–250 kDa (Bio-Rad, 1610373) and transferred onto a polyvinylidene fluoride (PVDF) membrane Trans-Blot Transfer Kit, (Bio-Rad, 1704272). To block non-specific binding, membranes were incubated for 2 h at room temperature in Tris-buffered saline with 0.1% Tween 20 (TBST, pH 7.4), supplemented with 5% dried skimmed milk. Following blocking, the membranes were incubated overnight at 4 °C under gentle shaking with CYR61 (E5W3H) Rabbit monoclonal primary antibodies (1:1,000; Cell Signaling, 39382S). After 24 h of incubation, membranes were washed with TBST and subsequently incubated with peroxidase (HRP) Anti-rabbit IgG goat secondary antibodies (Cell Signaling, 7074P) for 1 h at room temperature. Specific protein bands were visualized using Clarity Max Western ECL Substrate, (1705062, Bio-Rad laboratories). Membranes were stripped with Restore Western Blot Stripping Buffer, (Thermo Scientific, 21059), and reprobed for β-actin (13E5) rabbit monoclonal antibody (1:1,000; Cell Signaling, 4970S0). For quantitative analysis, protein band density was measured using ImageJ, with target signal normalization performed using the corresponding β-actin loading control. In the case of astrocyte conditioned media blots, protein loading was determined by Ponceau S staining.
Human multiple sclerosis and SCI spinal cord tissue
Human formalin-fixed paraffin-embedded (FFPE) spinal cord tissues from individuals with multiple sclerosis and neurologically healthy controls were prepared from autopsy-derived tissues collected by the rapid autopsy protocol approved by the Cleveland Clinic Institutional Review Board. Transverse spinal cord sections (7 μm) were prepared and the demyelinated lesions were identified by loss of proteolipid protein immunoreactivity. FFPE spinal cord tissues from individuals with SCI and associated clinical and neuropathological information were obtained from the International Spinal Cord Injury Biobank. The Clinical Research Ethics Board of the University of Columbia (Vancouver, Canada) granted permission for post-mortem spinal cord acquisition and for sharing biospecimens. Spinal cord biospecimens were collected from consented participants or their next of kin and provided as FFPE tissue sections at a thickness of 5 μm. SCI tissue sections evaluated herein derive from lesion-remote regions of the injured cord that exhibit white matter damage and/or Wallerian degeneration as determined by an experienced International Spinal Cord Injury Biobank neuropathologist on the basis of combined LFB with H&E staining, and results from 7 T magnetic resonance imaging of these spinal cord tissue blocks prior to sectioning. Deidentified information for healthy, multiple sclerosis and SCI patients is provided in Extended Data Fig. 9j.
Statistics, transparency and reproducibility
Statistical evaluations of repeated measures were performed using one-way or two-way ANOVA with post hoc independent pairwise analysis using Holm–Sidak test, Wilcoxon rank sum test, or t-tests using Prism 8 (GraphPad) unless indicated otherwise. In all cases, statistical measurements derive from means of biological replicates and error bars illustrate s.e.m. across biological replicates. P values are reported in the figures or figure legends. Differences with P < 0.05 were considered to be statistically significant. Power calculations were performed using G*Power Software v.3.1.9.2. All immunohistochemistry and in situ hybridization analyses shown were repeated at least three times with similar results. In graphs of histological continuous or count data, coloured data points represent the mean value for each biological replicate (individual mouse), while grey data points indicate replicate measurements from individual tissue sections. Mice were assigned numerical codes and randomized into experimental groups. In vitro culture experiments were repeated at least three times using independent cultures. Experimental procedures and quantitative analyses were conducted by individuals blinded to experimental group assignments.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.