This dataset was acquired, aligned and segmented as part of the larger MICrONS project. Methods underlying dataset acquisition are described in full detail elsewhere2,61,62,63, and the primary data resource is described in a separate publication2. We repeat some of the methodological details for the dataset here for convenience.
Animal preparation for EM
All animal procedures were approved by the Institutional Animal Care and Use Committee at the Allen Institute for Brain Science or Baylor College of Medicine. Neurophysiology data acquisition was conducted at Baylor College of Medicine before EM imaging. Afterwards the mice were transferred to the Allen Institute in Seattle and kept in a quarantine facility for 1–3 days, after which they were euthanized and perfused. All results described here are from a single male mouse, age 64 days at onset of experiments, expressing GCaMP6s in excitatory neurons via SLC17a7-Cre and Ai162 heterozygous transgenic lines (recommended and generously shared by Hongkui Zeng at the Allen Institute for Brain Science; JAX stock 023527 and 031562, respectively). Two-photon functional imaging took place between P75 and P80 followed by two-photon structural imaging of cell bodies and blood vessels at P80. The mouse was perfused at P87.
Tissue preparation
After optical imaging at Baylor College of Medicine, candidate mice were shipped via overnight air freight to the Allen Institute. Mice were transcardially perfused with a fixative mixture of 2.5% paraformaldehyde, 1.25% glutaraldehyde and 2 mM calcium chloride, in 0.08 M sodium cacodylate buffer, pH 7.4. A thick (1,200 µm) slice was cut with a vibratome and post-fixed in perfusate solution for 12–48 h. Slices were extensively washed and prepared for reduced osmium treatment based on the protocol of ref. 64. All steps were performed at room temperature, unless indicated otherwise. The first osmication step involved 2% osmium tetroxide (78 mM) with 8% v/v formamide (1.77 M) in 0.1 M sodium cacodylate buffer, pH 7.4, for 180 min. Potassium ferricyanide 2.5% (76 mM) in 0.1 M sodium cacodylate, 90 min, was then used to reduce the osmium. The second osmium step was at a concentration of 2% in 0.1 M sodium cacodylate for 150 min. Samples were washed with water and then immersed in thiocarbohydrazide for further intensification of the staining (1% thiocarbohydrazide (94 mM) in water, 40 °C, for 50 min). After washing with water, samples were immersed in a third osmium immersion of 2% in water for 90 min. After extensive washing in water, Walton’s lead aspartate (20 mM lead nitrate in 30 mM aspartate buffer, pH 5.5, 50 °C, 120 min) was used to enhance contrast. After two rounds of water wash steps, samples proceeded through a graded ethanol dehydration series (50%, 70%, 90% w/v in water, 30 min each at 4 °C, then 3 × 100%, 30 min each at room temperature). Two rounds of 100% acetonitrile (30 min each) served as a transitional solvent step before proceeding to epoxy resin (EMS Hard Plus). A progressive resin infiltration series (1:2 resin:acetonitrile (for eample, 33% v/v), 1:1 resin:acetonitrile (50% v/v), 2:1 resin acetonitrile (66% v/v) and then 2 × 100% resin, each step for 24 h or more, on a gyrotary shaker), was done before final embedding in 100% resin in small coffin moulds. Epoxy was cured at 60 °C for 96 h before unmoulding and mounting on microtome sample stubs. The sections were then collected at a nominal thickness of 40 nm using a modified ATUMtome (RMC/Boeckeler61) onto six reels of grid tape61,65.
Transmission EM imaging
The parallel imaging pipeline used in this study61 used a fleet of transmission electron microscopes that had been converted to continuous automated operation. It was built on a standard JEOL 1200EXII 120 kV transmission electron microscope that had been modified with customized hardware and software, including an extended column and a custom electron-sensitive scintillator. A single large-format CMOS (complementary metal–oxide–semiconductor) camera outfitted with a low-distortion lens was used to grab image frames at an average speed of 100 ms. The autoTEM was also equipped with a nano-positioning sample stage that offered fast, high-fidelity montaging of large tissue sections and a reel-to-reel tape translation system that locates each section using index barcodes. During imaging, the reel-to-reel GridStage moved the tape and located the targeting aperture through its barcode and acquired a 2D montage. We performed quality control on all image data and reimaged sections that failed the screening.
Image processing
Volume assembly
The volume assembly pipeline is described in detail elsewhere62,63. Briefly, the images collected by the autoTEMs are first corrected for lens distortion effects using nonlinear transformations computed from a set of 10 × 10 highly overlapping images collected at regular intervals. Overlapping image pairs are identified in each section, and point correspondences are extracted using features extracted using the scale-invariant feature transform. Montage transformation parameters are estimated per image to minimize the sum of squared distances between the point correspondences between these tile images, with regularization. A downsampled version of these stitched sections is produced for estimating a per-section transformation that roughly aligns these sections in three dimensions. The rough aligned volume is rendered to disk for further fine alignment. The software tool used to stitch and align the dataset is available on GitHub (https://github.com/AllenInstitute/render-modules). To fine align the volume, we needed to make the image processing pipeline robust to image and sample artefacts. Cracks larger than 30 um (in 34 sections) were corrected by manually defining transforms. The smaller and more numerous cracks and folds in the dataset were automatically identified using convolutional networks trained on manually labelled samples using 64 × 64 × 40 nm3 resolution images. The same was done to identify voxels containing tissue. The rough alignment was iteratively refined in a coarse-to-fine hierarchy66 using an approach based on a convolutional network to estimate displacements between a pair of images67. Displacement fields were estimated between pairs of neighbouring sections and then combined to produce a final displacement field for each image to further transform the image stack. Alignment was refined first using 1,024 × 1,024 × 40 nm3 images and then 64 × 64 × 40 nm3 images. The composite image of the partial sections was created using the tissue mask previously computed.
Segmentation
The image segmentation pipeline is fully described in ref. 62. Remaining misalignments were detected by cross-correlating patches of image in the same location between two sections after transforming into the frequency domain and applying a high-pass filter. Combining with the tissue map previously computed, a ‘segmentation output mask’ was generated that sets the output of later processing steps to zero in locations with poor alignment. Using previously described methods68, a convolutional network was trained to estimate intervoxel affinities that represent the potential for neuronal boundaries between adjacent image voxels. A convolutional network was also trained to perform a semantic segmentation of the image for neurite classifications, including (1) soma + nucleus, (2) axon, (3) dendrite, (4) glia and (5) blood vessel. Following the methods described in ref. 69, both networks were applied to the entire dataset at 8 × 8 × 40 nm3 in overlapping chunks to produce a consistent prediction of the affinity and neurite classification maps, and the segmentation output mask was applied to predictions. The affinity map was processed with a distributed watershed and clustering algorithm to produce an oversegmented image, where the watershed domains are agglomerated using single-linkage clustering with size thresholds70,71. The over-segmentation was then processed by a distributed mean affinity clustering algorithm70,71 to create the final segmentation.
For synapse detection and assignment, a convolutional network was trained to predict whether a given voxel participated in a synaptic cleft. Inference on the entire dataset was processed using the methods described in ref. 69 using 8 × 8 × 40 nm3 images. These synaptic cleft predictions were segmented using connected components, and components smaller than 40 voxels were removed. A separate network was trained to perform synaptic partner assignment by predicting the voxels of the synaptic partners given the synaptic cleft as an attentional signal72. This assignment network was run for each detected cleft, and coordinates of both the presynaptic and postsynaptic partner predictions were logged along with each cleft prediction.
For nucleus detection2, a convolutional network was trained to predict whether a voxel participated in a cell nucleus. Following the methods described in ref. 69, a nucleus prediction map was produced on the entire dataset at 64 × 64 × 40 nm3.
Column description and cell classes
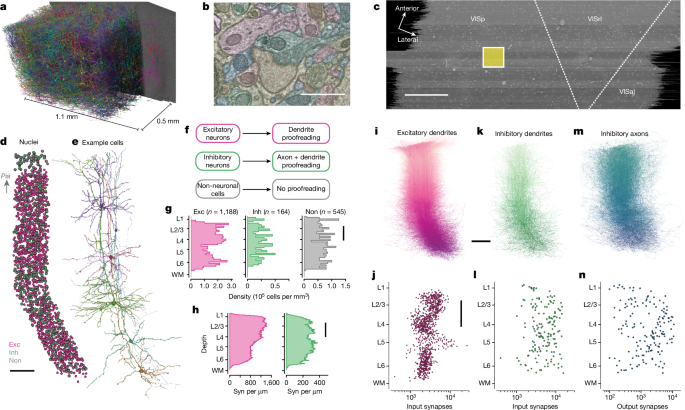
The column borders were found by manually identifying a region in primary visual cortex that was far from both dataset boundaries and the boundaries with higher-order visual areas. A 100 × 100 µm box was placed on the basis of layer 2/3 and was extended along the y axis of the dataset.
While analysing data, we observed that deep layer neurons had apical dendrites that were not oriented along the most direct pia-to-white-matter direction, and we adapted the definition of the column to accommodate these curved neuronal streamlines. Using a collection of layer 5 ET cells, we placed points along the apical dendrite to the cell body and then along the primary descending axon towards white matter. We computed the slant angle as two piecewise linear segments, one along the negative y axis to lower layer 5 where little slant was observed, and one along the direction defined by the vector-averaged direction of the labelled axons. We believe the slant to be a biological feature of the tissue and not a technical artefact for several reasons:
-
1.
The curvature is not aligned to a sectioning plane or associated with shearing or other distortion in the imagery, making it unlikely to be a result of the alignment process.
-
2.
Blood vessel segmentation does not show a large, correlated distortion in deep layers, making it unlikely to be a result of mechanical stress on the tissue (https://ngl.microns-explorer.org/#!gs://microns-static-links/mm3/blood_vessels.json). Moreover, it is unclear why such stress would affect only layer 5b and below.
-
3.
Individual examples of neurons with slanted morphologies can be found among single-cell reconstructions in the literature: for example, several descending bipolar VIP interneurons and layer 6 pyramidal cells in ref. 16. It is not possible to determine whether these individual cases correspond to a larger population of correlated arbours, but it suggests these morphologies are not atypical.
-
4.
Similar curvature has been observed in other large EM datasets from visual cortex (data not shown) and light level morphological reconstructions, particularly among layer 6 pyramidal cells.
Using these boundaries and previously computed nucleus centroids2, we identified all cells in the columnar volume. Coarse cell classes (excitatory, inhibitory and non-neuronal) were assigned on the basis of brief manual examination and rechecked by subsequent proofreading and automated cell typing40. To facilitate concurrent analysis and proofreading, we split all false merges connecting any column neurons to other cells (as defined by detected nuclei) before continuing with other work.
Proofreading
Proofreading was performed primarily by five expert neuroanatomists using the CAVE infrastructure73 and a modified version of Neuroglancer74. Proofreading was aided by on-demand highlighting of branch points and tips on user-defined regions of a neuron on the basis of rapid skeletonization (https://github.com/AllenInstitute/Guidebook). This approach quickly directed proofreader attention to potential false merges and locations for extension, as well as allowed a clear record of regions of an arbour that had been evaluated.
For dendrites, we checked all branch points for correctness and all tips to see if they could be extended. False merges of simple axon fragments onto dendrites were often not corrected in the raw data because they could be computationally filtered for analysis after skeletonization (see below). Detached spine heads were not comprehensively proofread, and previous estimates place the rate of detachment at roughly 10–15%. Using this method, dendrites could be proofread in about 10 min per cell.
For inhibitory axons, we began by ‘cleaning’ axons of false merges by looking at all branch points. We then performed extension of axonal tips until either their biological completion or data ambiguities, particularly emphasizing all thick branches or tips that were well-suited to project to new laminar regions. For axons with many thousands of synaptic outputs, we followed many but not all tips to completion once primary branches were cleaned and established. For smaller neurons, particularly those with bipolar or multipolar morphology, most tips were extended to the point of completion or ambiguity. Axon proofreading time differed significantly by cell type not only because of differential total axon length but also because of axon thickness differences that resulted in differential quality of autosegmentations, with thicker axons being of higher initial quality. Typically, inhibitory axon cleaning and extension took 3–10 h per neuron.
The lack of segmentation in the top 10 µm of layer 1 truncates some apical tufts and limited reconstruction quality of layer 1 interneurons. For those excitatory neurons with extensive apical tufts, particularly layer 2 and L5ET cells, the reconstructions here might miss both distinguishing characteristics and sources of inhibitory input in that region. Similarly, axons in deep layer 6 were generally less complete because of alignment quality in white matter.
Manual cell subclass and layer labels
Expert neuroanatomists further labelled excitatory and inhibitory neurons into subclasses. Layer definitions were based on considerations of both cell body density (in analogy with nuclear staining) supplemented by identifying kinks in the depth distribution of nucleus size near expected layer boundaries40.
For excitatory neurons, the categories used were Layer 2/3-IT, Layer 4-IT, Layer 5-IT, Layer 5-ET, Layer 5-NP, Layer 6-IT, Layer 6-CT and Layer 6b (‘L6-WM’) cells. Excitatory expert labels did not affect analysis but were used as the basis for naming morphological clusters. Layer 2/3 and upper Layer 4 cells were defined on the basis of dendritic morphology and cell body depth. Layer 5 cells were similarly defined by cell body depth, with projection subclasses distinguished by dendritic morphology following ref. 8 and classical descriptions of thick (ET) and thin-tufted (IT) cells. Layer 5 ET cells had thick apical dendrites, large cell bodies, numerous spines and a pronounced apical tuft, and deeper ET cells had many oblique dendrites. Layer 5 IT cells had more slender apical dendrites and smaller tufts, fewer spines and fewer dendritic branches overall. Layer 5 NP cells corresponded to the ‘Spiny 10’ subclass described in ref. 8; these cells had few basal dendritic branches, each very long and with few spines or intermediate branch points. Layer 6 neurons were defined by cell body depth, and some cells were able to be further labelled as IT or CT by human experts. Layer 6 pyramidal cells with stellate dendritic morphology, inverted apical dendrites or wide dendritic arbours were classified as IT cells. Layer 6 pyramidal cells with small and narrow basal dendrites, an apical dendrite ascending to Layer 4 or Layer 1 and a myelinated primary axon projecting into white matter were labelled as CT cells.
For inhibitory neurons, manual cell typing considered axonal and dendritic morphology as well as connectivity. Cells that primarily contacted soma or perisomatic regions were labelled as basket cells. Cells that made arbours that extended up to layer 1 or formed a dense plexus and primarily targeted distal dendrites were labelled as putative SST cells. Cells that remained mostly in layer 1 or had extensive arbourization and many non-synaptic boutons were labelled as putative Id2 or neurogliaform cells. Finally, cells with a bipolar dendritic morphology or a multipolar dendritic morphology and output onto other inhibitory neurons were labelled as putative VIP cells. Several cells, particularly in layer 6, had an ambiguous subclass assignment, typically when their connectivity was not basket-like but their morphology was also not similar to upper layer Martinotti or non-Martinotti cells.
Skeletonization
To rapidly skeletonize dynamic data, we took advantage of the PyChunkedGraph data structure that collects all supervoxels belonging to the same neuronal segmentation into 2 × 2 × 20 µm ‘chunks’ with a unique ID and precisely defined topological adjacency with neighbouring chunks of the same object. Each chunk is called a ‘level 2 chunk’, and the complete set of chunks for a neuron and their adjacency we call the ‘level 2 graph’ on the basis of its location in the hierarchy of the PyChunkedGraph data structure27. We precompute and cache a representative central point in space and the volume and the surface area for each level 2 chunk and update this data when new chunks are created because of proofreading edits. Using the level 2 graph and assigning edge lengths corresponding to the distance between the representative points for each vertex (that is, each level 2 chunk), we run the TEASAR75 algorithm (10 µm invalidation radius) to extract a loop-free skeleton. Each of the level 2 vertices removed by the TEASAR algorithm is associated with its closest remaining skeleton, making it possible to map surface area and volume data to the skeleton. Typical edges between skeleton vertices are about 1.7 µm, and new skeletons can be computed de novo in about 10 s, making them useful for analysis over length scales of tens of micrometre or larger.
To represent the cell body, a further vertex was placed at the location of the nucleus centroid, and all vertices within an initial radius and topologically connected to centroid were collapsed into this vertex with associated data mapping. The radius was determined for each neuron separately by consideration of the volume of each cell body. A companion work40 computed the volume of each cell body, and we generated an effective radius on the basis of the sphere with the same volume. To ensure that our values captured potentially lopsided cell bodies, we padded this effective radius by a further factor of 1.25. Skeletons were rooted at the cell body, with ‘downstream’ meaning away from soma and ‘upstream’ meaning towards soma. Each synapse was assigned to skeleton vertices on the basis of the level 2 chunk of its associated supervoxel. For each unbranched segment of the skeleton (that is, between two branch points or between a branch point and end point), we computed an approximate radius r on the basis of a cylinder with the same path length L and total volume V associated with that segment: \((r=\sqrt{V/{\rm{\pi }}L})\).
Axon/dendrite classification
To detect axons, we took advantage of the skeleton morphology, the location of presynaptic and postsynaptic synapses and the clear segregation between inputs and outputs of cortical neurons. For inhibitory cells, we used synapse flow centrality76 to identify the start of the axon as the location of maximum paths along the skeleton between sites of synaptic input and output. Two inhibitory neurons had two distinct, biologically correct axons after proofreading (cell IDs 258362 and 307059). For these cells, we ran this method twice, masking off the axon found after the first run, to identify both. For excitatory neurons that did not have extended axons, there were often insufficient synaptic outputs on their axon for this approach to be reliable. Excitatory neurons with a segregation index76 of 0.7 (on a scale with 0 indicating random distribution of input and output synapses and 1 indicating perfect input/output segregation) or above were considered well-separated, and the synapse flow centrality solution was used. For cells with a segregation index less than 0.7, we instead looked for branches near the soma with few synaptic inputs. Specifically, we took identified all skeleton vertices within 30 µm of the cell body and looked at the distinct branches downstream from this region. For each branch, we computed the total path length and the total number of synaptic inputs to get a linear input density. Branches with both a path length more than 20 µm and an input density less than 0.1 synaptic inputs per micrometre were labelled as being axonal and filtered out of subsequent analysis.
We further filtered out any remaining axon fragments merged onto pyramidal cell dendrites using a similar approach. We identified all unbranched segments (regions between two branch points or between a branch point and end point) on the non-axonal region of the skeleton and computed their input synapse density. Starting from terminal segments (that is, those with no downstream segments), we labelled a segment as a ‘false merge’ if it had an input density less than 0.1 synaptic inputs per micrometre. This process iterated across terminal segments until all remaining had an input density of at least 0.1 inputs per micrometre. Falsely merged segments were masked out of the skeleton for all analysis.
Excitatory dendrite compartments
We assigned all synaptic inputs onto excitatory neurons to one of four compartments: soma, proximal dendrite, distal basal dendrite and distal apical dendrite. The most complex part was distinguishing the basal dendrite from the apical dendrite. Although easy in most cases for neurons in layer 3–5 because of the consistent nature of apical dendrites being single branches reaching towards layer 1, this is not true everywhere. In upper layer 2/3, cells often have several branches in layer 1 equally consistent with apical dendrites, and in layer 6 there are often cells with apical dendrites that stop in layer 4 and that point towards white matter or even that lack a clear apical branch entirely. To objectively and scalably define apical dendrites, we built a classifier that could detect between zero and three distinct apical branches per cell. Following the intuition from neuroanatomical experts, we used features on the basis of the branch orientation, location in space, relative location compared to the cell body and branch-level complexity. Specifically, we trained a random forest classifier to predict whether a skeleton vertex belonged to an apical dendrite on the basis of several features: depth of vertex, depth of soma, difference in depth between soma and vertex, vertex distance to soma along the skeleton, vertex distance to farthest tip, normalized vertex distance to tip (between 0 and 1), tortuosity of path to root, number of branch points along the path to root, radial distance from soma, absolute distance from soma and angle relative to vertical between the vector from soma to vertex. We aggregated predictions in each branch by summing the log-odds ratio from the model prediction, with the net log-odds ratio saturating at ±200. Finally, for each branch i with aggregated odds ratio Ri, we compare branches to one another via a soft-max operation: \({S}_{i}=\exp \left({R}_{i}/50\right)/\,{\sum }_{j}\exp ({R}_{j}/50)\). Branches with a maximum tip length of less than 50 µm were considered too short to be a potential apical dendrite and excluded from consideration and not included in the denominator. Branches with both Ri > 0 (evidence is positive towards being apical) and Si > 0.25 were defined to be apical. Note that the soft-max was chosen to allow multiple apical branches if they had similar aggregated odds ratios, which was found to be necessary for upper layer pyramidal neurons. Training data were selected from an initial 50 random cells, followed by a further 33 cells chosen representing cases where the classifier did not perform correctly. Performance on both random and difficult cells had an F1-score of 0.9297 (86 true positives, 599 true negatives, 2 false positives and 11 false negatives) on the basis of leave-one-out cross validation, with at least one apical dendrite correctly classified for all cells.
Compartment labels were propagated to synapses on the basis of the associated skeleton vertices. Soma synapses were all those associated with level 2 chunks in the soma collapse region (see the section ‘Skeletonization’). Proximal dendrites were those outside of the soma but within 50 µm after the start of the branch. Distal basal synapses were all those associated with vertices more distant than the proximal threshold but not on an apical branch. Apical synapses were all those associated with vertices more distant than the proximal threshold and on an apical branch.
Inhibitory feature extraction and clustering
Many classical methods of distinguishing interneuron classes are based on how cells distribute their synapses across target compartments. Following proofreading, expert neuroanatomists attempted to classify all inhibitory neurons broadly as ‘basket cells’, ‘SST-like cells’, ‘VIP-like cells’ and ‘neurogliaform/layer 1’ cells on the basis of connectivity properties and morphology. Although 150 cells were labelled on this basis, a further 13 neurons were considered uncertain (primarily in layer 6), and in some cases manual labels were low confidence. To classify inhibitory neurons in a data-driven manner, we thus measured four properties of how cells distribute their synaptic outputs:
-
1.
The fraction of synapses onto inhibitory neurons.
-
2.
The fraction of synapses onto excitatory neurons that are onto soma.
-
3.
The fraction of synapses onto excitatory neurons that are onto proximal dendrites.
-
4.
The fraction of synapses onto excitatory neurons that are onto distal apical dendrites.
Because the fraction of synapses targeting all compartments sums to one, the last remaining property, synapses onto distal basal dendrites, was not independent and thus was measured but not included as a feature. Inspection of the data suggested two more properties that characterized synaptic output across inhibitory neurons:
-
5.
The fraction of synapses that are part of multisynaptic connections, those with at least two synapses between the same presynaptic neuron and target neuron.
-
6.
The fraction of multisynaptic connection synapses that were also within 15 µm of another synapse with the same target, as measured between skeleton nodes. Note that we evaluated the robustness of this parameter and found that intersynapse distances from 5 to more than 100 µm have qualitatively similar results (Extended Data Fig. 2).
Using these six features, we trained a linear discriminant classifier on cells with manual annotations and applied it to all inhibitory cells. Differences from manual annotations were treated not as inaccurate classifications but rather as a different view of the data.
Excitatory feature extraction and clustering
To characterize excitatory neuron morphology, we computed features based only on excitatory neuron dendrites and soma. The features were as follows:
-
1.
Median distance from branch tips to soma per cell.
-
2.
Median tortuosity of the path from branch tips to soma per cell. Tortuosity is measured as the ratio of path length to the Euclidean distance from tip to soma centroid.
-
3.
Number of synaptic inputs on the dendrite.
-
4.
Number of synaptic inputs on the soma.
-
5.
Net path length across all dendritic branches.
-
6.
Radial extent of dendritic arbour. We define ‘radial distance’ to be the distance in the same plane as the pial surface. For every neuron, we computed a pia-to-white-matter line, including slanted region in deep layers, passing through its cell body. For each skeleton vertex, we computed the radial distance to the pia-to-white-matter line at the same depth. To avoid any outliers, the radial extent of the neuron was defined to be the 97th percentile distance across all vertices.
-
7.
Median distance to soma across all synaptic inputs.
-
8.
Median synapse size of synaptic inputs onto the soma.
-
9.
Median synapse size of synaptic inputs onto the dendrites.
-
10.
Dynamic range of synapse size of dendrite synaptic inputs. This was measured as the difference between 95th and fifth percentile synapse sizes.
-
11.
Shallowest extent of synapses, on the basis of the fifth percentile of synapse depths.
-
12.
Deepest extent of synapses, on the basis of the 95th percentile of synapse depths.
-
13.
Vertical extent of synapses, on the basis of the difference between 95th and fifth percentile of synapse depths.
-
14.
Median linear density of synapses. This was measured by computing the net path length and number of synapses along 50 depth bins from layer 1 to white matter and computing the median. A linear density was found by dividing synapse count by path length per bin, and the median was found across all bins with non-zero path length.
-
15.
Median radius across dendritic skeleton vertices. To avoid the region immediately around the soma from having a potential outlier effect, we only considered skeleton vertices at least 30 µm from the soma.
Three more sets of features used component decompositions. To more fully characterize the absolute depth distribution of synaptic inputs, for each excitatory neuron, we computed the number of synapses in each of 50 depth bins from the top of layer 1 to surface of white matter (bin width approximately 20 µm). We Z-scored synapse counts for each cell and computed the top six components using SparsePCA. The loadings for each of these components on the basis of the net synapse distribution were used as features.
To characterize the distribution of synaptic inputs relative to the cell body instead of cortical space, we computed the number of synapses in 13 soma-adjusted depth bins starting 100 µm above and below the soma. As before, synapse counts were Z-scored, and we computed the top five components using SparsePCA. The loadings for each of these components were used as further features.
To characterize the relationship with branching to distance, we measured the number of distinct branches as a function of distance from the soma at ten distances, every 30 µm starting at 30 µm from the soma and continuing to 300 µm. For robustness relative to precise branch point locations, the number of branches were computed by finding the number of distinct connected components of the skeleton found in the subgraph formed by the collection of vertices between each distance value and 10 µm towards the soma. We computed the top three singular value components of the matrix on the basis of branch count versus distance for all excitatory neurons, and the loadings were used as features.
All features were computed after a rigid rotation of 5 degrees to flatten the pial surface and translation to set the pial surface to 0 on the y axis. Features on the basis of apical classification were not explicitly used to avoid ambiguities on the basis of both biology and classification.
Using this collection of features, we clustered excitatory neurons by running phenograph77 500 times with 95% of cells included each time. Phenograph finds a nearest-neighbourhood graph on the basis of proximity in the feature space and clusters by running the Leiden algorithm for community detection on the graph. Here we used a graph on the basis of ten nearest neighbours and clustered with a resolution parameter of 1.3. These values were chosen to consistently separate layer 5 ET, IT and NP cells from one another, a well-established biological distinction. A coclustering matrix was assembled with each element corresponding to the number of times two cells were placed in the same cluster. To compute the final consensus clusters, we performed agglomerative clustering with complete linkage on the basis of the coclustering matrix, with the target number of clusters set by a minimum Davies–Bouldin score and a maximum Silhouette score. Clusters were then named on the basis of the most frequent manually defined cell type in the cluster and reordered on the basis of median soma depth. The labelling of cells as layer 2 and layer 3 was formed on the basis of soma depth and a morphology with a relatively flat morphology, often with no distinct apical trunk, although often apical-tuft-like branches emitted directly from the cell body. The L2c subclass was ambiguously defined between the two categories, with cells that had a distinct apical trunk but with connectivity and other properties that seemed more similar to layer 2 subclasses.
To compute the importance of each feature for each M-type, for each M-type we trained a random forest classifier to predict whether a cell belonged to it using scikit-learn78. Because the classes were strongly imbalanced, we used SMOTE resampling to oversample datapoints from the smaller class. We used the Mean Decrease in Impurity metric, which quantifies how often a given feature was used in the decision tree ensemble.
Inhibitory connectivity and selectivity
To measure intracolumnar inhibitory connectivity, we first restricted synaptic outputs to the axon of each inhibitory neuron, as we have not observed any correctly classified synaptic outputs on dendritic arbours in this dataset. One cell with fewer than 30 synaptic outputs was omitted because of insufficient size. All remaining synaptic outputs across all interneurons were then filtered to include only those that target cells in the column, unless otherwise specified. Each output synapse was also labelled with the target skeleton vertex, dendritic compartment and M-type of the target neuron on the basis of the compartment definitions above.
For the inhibitory motif group clustering, for each interneuron we first computed the number of synapses across each excitatory M-types in the column. This synaptic output budget was then normalized per cell to generate a vector for each neuron with elements ranging from zero to one. Normalized synaptic output budgets were oversegmented using k-means (k = 20) with Euclidean distances 500 times, and a matrix of coclustering frequency—that is, the number of times two cells were put in the same k-means cluster—between individual cells was computed. Final M-types were found through agglomerative clustering with complete linkage of the coclustering matrix, scanning from two to 25 output clusters and selecting a final value of 18 on the basis of silhouette score and Davies–Bouldin score.
For measuring the synaptic output budget across cell types across the dataset (that is, inside and outside the column), we used a hierarchical classifier on the basis of a collection of perisomatic features that was trained on the data-driven clustering from the column sample40. Only synapses onto object segmentation associated with a single nucleus and a cell type classification were used. Although most of these other targets were not proofread, estimates on the basis of proofread neurons indicate that 99% of non-proofread input synapses are accurate40.
To measure inhibitory selectivity in the column, we compared the M-type distribution of its synaptic outputs to the M-type distribution of synaptic inputs according to a null model accounting for cell abundance, synapse abundance and depth. We first generated a baseline distribution of all 4,504,935 somatic or dendritic synaptic inputs to all column cells, where each synapse was associated with a precise depth, target compartment and M-type. We discretized synaptic inputs into 50 depth bins spanning pia to white matter, each covering approximately 20 µm and each of the five compartments: soma, proximal dendrite, basal dendrite, apical dendrite or inhibitory neuron. For each interneuron, we similarly discretized its synaptic output into the same bins, compartments and M-types. To generate a randomized output distribution preserving both observed depth and compartment distributions, we randomly picked synapses from the baseline distribution with the observed depth bins and compartment targets but without regard to M-type. We computed 10,000 randomized distributions per interneuron. To get a selectivity index, we compared the observed number of synapses onto a given M-type to the median of the number of synapses from the shuffle distribution. To get a significance for the selectivity index for a given M-type, we directly computed the two-sided P value of the observed number of synapses relative to the shuffle distribution for that M-type. P values were corrected for several comparisons using the Holm–Sidak method in each interneuron for those M-types with non-zero potential connectivity. Selectivity was only measured in the column because we did not generate compartment labels for unproofread dendrites outside of the column.
On connectivity cards, we also show a similar selectivity index on the basis of compartment rather than M-type. In that case, the shuffled distribution preserves observed depth and M-type output distributions but not compartments.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.