Generation of knock-in mice by CRISPR–Cas9 targeting
The Spo11D277N mouse line was generated by Shanghai Biomodel Organism using CRISPR–Cas9 technology. The donor oligos were designed to change the D277 codon GAT (D) to AAT (N). The resulting point-mutation founders were back-crossed with C57BL/6J mice to obtain heterozygous mutant mice. The heterozygous mutants were crossbred to generate homozygous Spo11D277N mutant mice. Genotyping of Spo11D277N was done by PCR and DNA sequencing. The sequences of injected sgRNA, oligo donors and primers for genotyping are shown in Supplementary Table 1. Mice were fed regular rodent chow with ad libitum access to water and food in a 12-h light–dark cycle in a temperature and humidity-controlled environment (20–26 °C, 30–70% humidity). All animal experiments were performed according to the guidelines of the Animal Care and Use Committee at Shanghai Institute of Biochemistry and Cell Biology, Center for Excellence in Molecular Cell Science, Chinese Academy of Sciences. The experiments performed in this study were approved by the Ethics Committee of Center for Excellence in Molecular Cell Science, Chinese Academy of Sciences (2022-151).
Preparation of expression vectors
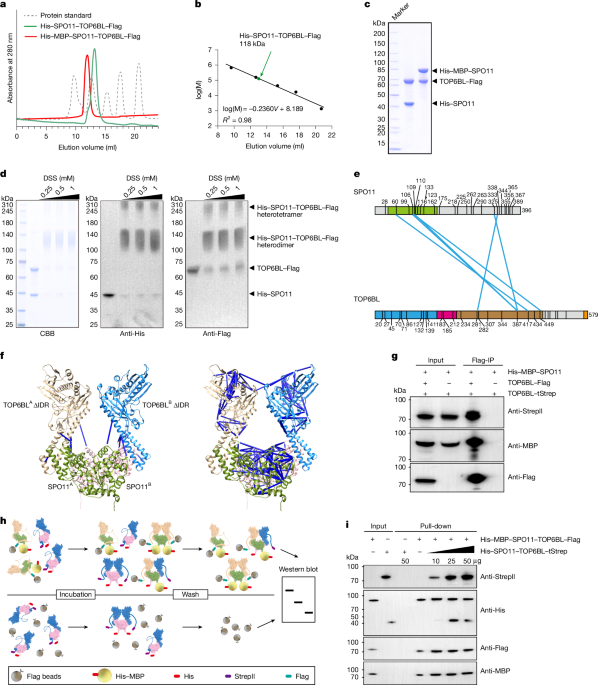
Expression vectors for N-terminally His–MBP-tagged SPO11 and C-terminally Flag-tagged TOP6BL were generated by cloning sequences derived from codon-optimized, synthesized mouse Spo11 and Top6bl cDNA (Genewiz) into the pcDNA3.4-MBP vector and the pMC vector (a gift from J. Liu), respectively. To construct a version of TOP6BL with a C-terminal Twin-Strep tag (TOP6BL–tStrep), the Top6bl coding sequence was cloned into a pMC vector that had been modified to include the twin StrepII tag sequence at the 3′ end. To obtain SPO11 without the MBP tag, the pcDNA3.4-His-MBP-SPO11 construct was PCR-amplified using the SPO11_FO/RO primer pairs and self-ligated using MultiF Seamless Assembly Mix (ABclonal), according to the manufacturer’s instructions. Point mutants were also generated using MultiF Seamless Assembly Mix (ABclonal). The sequences of the above-mentioned plasmid are shown in Supplementary Fig. 2. For plasmid coding for human TDP2, a DNA fragment encoding the full length of human TDP2 was cloned into the pET28-SMT3 vector between the BamHI and XhoI sites, which contains an N-terminal Ulp1-cleavable His6–SUMO tag. The oligonucleotides for this study were synthesized by Sangon, and the plasmids used in this study are listed in Supplementary Tables 2 and 3.
Expression and purification of the mouse SPO11–TOP6BL complex
Three variants of mouse SPO11–TOP6BL complexes (His–SPO11–TOP6BL–Flag, His–MBP–SPO11–TOP6BL–Flag and His–SPO11–TOP6BL–tStrep) were expressed in Expi293F cells. For each complex, plasmids were co-transfected at a 1:1 mass ratio by 2 mg plasmids in 1 l culture. Cells were cultured in 4 l suspension at 37 °C with 5% CO2 and 125 rpm orbital shaking. After 72 h, cells were collected (500g, 5 min) and lysed in buffer containing 25 mM HEPES, pH 7.5, 150 mM NaCl and 20 mM imidazole, 10% glycerol, 0.5% Triton X-100, 1 mM DTT and 1× protease inhibitor cocktail (Sparkjade) using a high-pressure crusher (Union, 300–500 Pa, 5 min). The lysate was clarified by centrifugation (48,000g, 90 min).
All three complexes were initially purified using nickel affinity chromatography. The supernatant was incubated with 2 ml pre-equilibrated Ni-NTA resin (QIAGEN) for 2 h. After washing with 200 ml buffer (25 mM HEPES, pH 7.5, 150 mM NaCl, 40 mM imidazole and 10% glycerol), proteins were eluted with 50 ml buffer containing 25 mM HEPES, pH 7.5, 150 mM NaCl, 300 mM imidazole, 10% glycerol, 1 mM DTT and 1× protease inhibitor cocktail.
Then, the eluate was supplemented with 10 mM EDTA and incubated with 1 ml pre-equilibrated Flag resin (Selleckchem) for 2 h. After washing with 50 ml buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 10% glycerol), proteins were eluted with 5 ml buffer containing 50 mM Tris, pH 7.5, 150 mM NaCl, 10% glycerol, 0.1 mg ml−1 3× Flag peptide (MCE), 1 mM DTT and 1× protease inhibitor cocktail.
For tStrep-tagged complex, the Ni-NTA eluate was incubated with 1 ml pre-equilibrated StrepTactin XT resin (IBA) for 2 h. After washing with 50 ml buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 10% glycerol), proteins were eluted with 5 ml buffer containing 50 mM Tris, pH 7.5, 150 mM NaCl, 10% glycerol, 2.5 mM desthiobiotin, 1 mM DTT and 1× protease inhibitor cocktail.
All purified complexes were further fractionated by SEC using a Superdex 200 Increase 10/300 GL column (Cytiva) equilibrated in 20 mM HEPES, pH 7.5, 100 mM NaCl and 0.25 mM TCEP. The column was calibrated using Bio-Rad’s Gel Filtration Standard (1511901). Protein concentrations were determined by Bradford assay (Thermo Fisher Scientific).
Co-immunoprecipitation
Expi293F cells were cultured in suspension and split into two 100-ml cultures. When the cell density reached approximately 3 × 106 cells per ml, one culture was transfected with plasmids encoding His–MBP–SPO11 (0.2 mg), TOP6BL–Flag (0.1 mg) and TOP6BL–tStrep (0.1 mg). The second culture was transfected with plasmids encoding His–MBP–SPO11 (0.1 mg) and TOP6BL–tStrep (0.1 mg). After 72 h, cells were collected by centrifugation at 500g for 5 min at 4 °C and resuspended in 25 ml lysis buffer containing 20 mM HEPES, pH 7.5, 100 mM NaCl, 0.25 mM Tris(2-carboxyethyl)phosphine (TCEP) and 1× protease inhibitor cocktail. Cells were lysed by gentle sonication on ice for 1 h. The lysates were clarified by centrifugation at 48,000g for 90 min at 4 °C.
For each sample, 10 ml of supernatant was incubated overnight at 4 °C with 50 μl of pre-equilibrated anti-Flag M2 affinity gel beads. After incubation, the beads were collected by centrifugation at 500g for 5 min at 4 °C and washed three times with 2 ml of wash buffer (20 mM HEPES, pH 7.5, 100 mM NaCl, 0.25 mM TCEP, 0.1% NP-40 and 1× protease inhibitor cocktail). Bound proteins were eluted by incubating the beads with 100 μl of elution buffer containing 20 mM HEPES, pH 7.5, 100 mM NaCl, 0.25 mM TCEP and 0.1 mg ml−1 Flag peptide for 1 h at 4 °C with gentle agitation.
Eluates were mixed with SDS–PAGE loading buffer, boiled at 95 °C for 5 min and analysed by western blotting using standard protocols. The tStrep tag was detected with StrepII tag mouse monoclonal antibody (HRP conjugated, AF2927, Beyotime), the Flag tag was detected with Flag tag mouse monoclonal antibody (HRP conjugated, AF2855-50μl, Beyotime) and the MBP tag was detected with MBP tag mouse monoclonal antibody (HRP conjugated, AF2915, Beyotime).
Pull-down assay
Fifty microlitres of anti-Flag M2 affinity gel beads were equilibrated with a binding buffer containing 20 mM HEPES, pH 7.5, 100 mM NaCl and 0.25 mM TCEP. In a total volume of 200 µl of binding buffer, 5 µg of purified His–MBP–SPO11–TOP6BL–Flag was mixed with varying amounts of His–SPO11–TOP6BL–Strep (0, 10, 25 or 50 μg) and added to the equilibrated Flag beads for incubation at 4 °C for 3 h with gentle rotation. After centrifugation at 500g for 5 min at 4 °C, the supernatant was removed, and the beads were washed three times with 1 ml binding buffer to eliminate unbound proteins. The beads were then resuspended in 50 µl elution buffer (20 mM HEPES, pH 7.5, 100 mM NaCl, 0.25 mM TCEP and 0.1 mg ml−1 Flag peptide) and incubated at 4 °C for 2 h with gentle agitation to elute bound proteins. The eluted proteins were analysed by western blotting, detecting the His tag with His tag mouse monoclonal antibody (HRP conjugated, AF2879-50μl, Beyotime). The remaining antibodies used for detecting the MBP, Flag and tStrep tags were the same as those used in the co-immunoprecipitation experiments.
Expression and purification of human TDP2
To express recombinant proteins, the plasmid containing the cDNA of full-length human TDP2 was transformed into Escherichia coli BL21(DE3). Cells were cultured in LB medium at 37 °C with 50 mg ml−1 kanamycin until the optical density at 600 nm reached 0.6–0.8. The cells were cooled at 18 °C for one hour before induction with 0.2 mM IPTG for 16 h. Cells were collected by centrifugation at 5,000 rpm for 10 min. Cell pellets were resuspended in lysis buffer containing 20 mM Tris, pH 8.0, 500 mM NaCl and 25 mM imidazole and lysed using a high-pressure homogenizer (Litu, FB-110×15). The cell lysates were centrifuged at 16,000 rpm for 1 h at 4 °C. The supernatants were then purified using a nickel-chelating affinity column (HisTrap, Cytiva), washed with lysis buffer and eluted with a buffer containing 20 mM Tris, pH 8.0, 500 mM NaCl and 500 mM imidazole. The target fusion protein was incubated with Ulp1 protease to remove the SUMO tag. The mixture was diluted fivefold with 20 mM Tris, pH 8.0 and loaded onto a HiTrap Q HP column (Cytiva) to remove the SUMO tag and Ulp1 protease. The eluate was concentrated onto a HiLoad 16/60 Superdex 200 column (Cytiva) equilibrated with a buffer containing 20 mM HEPES, pH 7.5 and 300 mM NaCl. The purified proteins were then concentrated and stored at −80 °C for subsequent use.
DNA-cleavage assays
For plasmid DNA-cleavage assays, a 4,000-bp T-vector (pEASY-Blunt, Trans) containing a SPO11 hotspot sequence (GRCm38p2/mm10 chr. 19: 59349669–59349759) was used as the substrate (hereafter referred to as pEBZSH)21. The reactions were conducted in 10 μl cleavage buffer containing 50 mM HEPES, pH 7.5, 30 mM KCl, 2 mM DTT and 1 mM MgCl2, unless otherwise specified. Protein complexes and the plasmid substrate were initially prepared on ice.
Time-course experiments were performed to assess the kinetics of cleavage, using 400 nM concentrations of His–SPO11–TOP6BL–Flag, His–MBP–SPO11–TOP6BL–Flag and His–MBP–SPO11(Y138F)–TOP6BL–Flag. Each reaction contained 5 nM plasmid substrate and was incubated at 37 °C, with samples collected at intervals of 0, 5, 10, 15, 30, 60 and 90 min.
For mutational analysis, the catalytic activities of wild-type SPO11 and the mutants Y137F, Y138F and Y137F/Y138F were compared across protein concentrations of 50, 100, 200 and 400 nM. Metal-binding site mutants (E224Q, D277N and D279N) were analysed under the same concentration conditions.
To investigate metal-ion dependence, reactions included varying the concentrations of MgCl2, MnCl2 or CaCl2, ranging from 0 to 20 mM. Nucleotide requirements were tested by supplementing with 0 to 5 mM ATP or other NTPs or dNTPs, with a constant protein concentration of 400 nM.
Substrate and protein titration experiments were designed to investigate the concentration effects. The effect of substrate concentration was evaluated at a constant protein concentration of 400 nM, with plasmid levels set between 1.25 and 20 nM. Conversely, protein concentration effects were assessed at fixed plasmid concentrations (1.25 nM and 20 nM), with protein titrated from 0 nM to 400 nM.
For linear DNA substrate preparation, the plasmid was digested by EcoRI (NEB) and purified with the FastPure Gel DNA extraction kit (Vazyme). Cleavage assays were performed with 400 nM of wild-type protein under two conditions: one adhering to standard buffer conditions, and the other substituting Mn2+ for Mg2+ while omitting KCl.
Reactions were terminated by adding 1 μl 10% SDS, 0.5 μl 500 mM EDTA and 0.8 μl proteinase K (Roche, 20 mg ml−1), followed by incubation at 55 °C for 30 min. Reaction products were analysed by electrophoresis on 1% agarose gels in TAE buffer at 80 V for 60 min. Gels were stained with GelRed (Yeasen) and visualized using a Tanon 2500 imaging system. For each plasmid cleavage assay, an equal amount of linear DNA was included as a positive control. The production of linear DNA was quantified using ImageJ software.
Identification of the 5′ tyrosine–phosphate bond in SPO11–DNA
The SPO11 cleavage reactions were performed as described previously for the DNA-cleavage assays, using 400 nM His–MBP–SPO11–TOP6BL–Flag complex incubated with plasmid DNA at 37 °C for 30 min. The cleavage buffer consisted of 50 mM HEPES, pH 7.5, 30 mM KCl, 2 mM DTT and 1 mM MgCl2.
To assess the formation of protein–DNA conjugates, reactions containing wild-type, Y137F or Y138F His–MBP–SPO11–TOP6BL–Flag complexes were processed with or without 1% SDS and proteinase K (Roche, 20 mg ml−1). The samples were then loaded onto 1% agarose gels and subjected to electrophoresis in TAE buffer. After electrophoresis, the gels were stained with GelRed (Yeasen) for visualizing DNA bands.
For the immunoprecipitation assay, wild-type and Y138F complexes were reacted with plasmid DNA (total 500 μl). After the reaction, product was supplemented with 50 mM HEPES 7.5, 4 M urea and 250 mM NaCl, and then incubated with 25 μl pre-equilibrated Ni-NTA resin (QIAGEN). The beads were washed three times with 1 ml wash buffer (50 mM HEPES 7.5, 4 M urea and 250 mM NaCl). Bound complexes were eluted by incubating the beads with 25 μl elution buffer containing 1% SDS in 25 mM HEPES, pH 7.5, 150 mM NaCl and 10% glycerol for 10 min at room temperature with gentle mixing. The isolated complexes were treated with proteinase K, followed by analysis on agarose gels to check for the presence of DNA bands indicative of cleavage.
To examine the role of human TDP2 in cleaving the 5′ tyrosine–phosphate bond, wild-type and Y138F complexes were reacted with plasmid DNA. Reactions were terminated with a brief heating at 70 °C for 10 min to denature the SPO11–TOP6BL complex. Human TDP2 (800 nM) was added to the reactions, which were subsequently incubated at 37 °C for 30 min. Reactions were terminated with 1 µl of 10% SDS and 0.5 µl of 500 mM EDTA. The reaction products, with or without proteinase K and human TDP2 treatment, were analysed on agarose gels.
A 46-bp oligonucleotide substrate labelled with Cy3 fluorescence at either the 5′ or 3′ end was used to evaluate covalent-bond formation with SPO11. The sequence is SPO11 hotspot (GRCm38p2/mm10 chr. 6: 93618016-93618061)21. Reaction mixtures (10 μl total volume) contained 0.5 μM Cy3-labelled oligonucleotide and 2 μM of either wild-type or Y138F-mutant His–MBP–SPO11–TOP6BL–Flag complexes. The reactions were performed in buffer comprising 20 mM HEPES, pH 7.5, 30 mM KCl, 2 mM MgCl2, 2 mM MnCl2 and 5% (v/v) glycerol. After the cleavage reaction, Flag-tagged proteins and associated oligonucleotides were captured using anti-Flag affinity beads. The reaction mixtures were incubated with pre-equilibrated anti-Flag beads (50 μl) at 4 °C with gentle agitation for one hour in wash buffer containing 20 mM HEPES, pH 7.5, 100 mM NaCl, 0.1% NP-40, 0.25 mM TCEP and 1 mM EDTA. After binding, the beads were washed three times with the wash buffer to remove non-specifically bound materials. Each wash involved gently resuspending the beads for 15 min at 4 °C, followed by centrifugation at 500g for 2 min. Bound complexes were eluted by incubating the beads with 20 μl elution buffer containing 0.1 mg ml−1 Flag peptide in 20 mM HEPES, pH 7.5, 100 mM NaCl and 0.25 mM TCEP for 30 min at 4 °C with gentle mixing. Eluted products were analysed by SDS–PAGE, and fluorescence was detected using a Typhoon FLA 9000 gel imaging system (Cytiva) capable of detecting Cy3 fluorescence. For further analysis of reaction products, further reactions were performed at varying concentrations of His–MBP–SPO11–TOP6BL–Flag complexes. Reactions were terminated by adding SDS to a final concentration of 1% and EDTA to 50 mM (pH 8.0). Each sample was treated with 100 μg of proteinase K. All samples were subjected to 8 M urea–PAGE to separate cleavage fragments. The oligonucleotides (GenScript) used in this study are listed in Supplementary Table 5.
EMSA
The sequence of the PRDM9 hotspot motif was used for this study21. Purified His–MBP–SPO11–TOP6BL–Flag was incubated with Cy3-labelled DNA oligos at a final concentration of 10 nM in a binding buffer containing 20 mM HEPES, pH 7.5 and 30 mM KCl for 30 min at 4 °C. After incubation, bound and free DNA probes were separated by 6% native PAGE conducted at 4 °C. Cy3 fluorescence was visualized using a Typhoon FLA 9000 gel imaging system. The DNA-binding ratios were quantified by measuring the reduction in free DNA, with the grey value of free DNA quantified using ImageJ software. Oligonucleotides (GenScript) used in this study are listed in Supplementary Table 5.
XL-MS
The purified His–SPO11–TOP6BL–Flag complex was incubated with 0.25 mM, 0.5 mM or 1 mM DSS (Pierce, A39267) in a reaction buffer comprising 20 mM HEPES, pH 7.5 and 30 mM KCl at 25 °C for 30 min. The reaction was terminated by adding quenching buffer (50 mM Tris, pH 8.0) to a final concentration of 20 mM, followed by a 20-min incubation at room temperature. One-third of the sample was analysed using 10% SDS–PAGE and Coomassie blue staining to assess cross-linking efficiency. The remaining sample was precipitated with acetone. The protein pellet was dried using a Speedvac for 1–2 min. The pellet was subsequently dissolved in 8 M urea, 100 mM Tris, pH 8.5 and 5 mM TCEP (Thermo Fisher Scientific), and 10 mM iodoacetamide (Sigma) for reduction and alkylation was added to the solution, followed by incubation at room temperature for 30 min, respectively. The protein mixture was diluted four times and digested overnight with trypsin at 1:50 (w/w) (Promega). The digested peptide solutions were desalted using a MonoSpin C18 column (GL Science) and dried with a SpeedVac.
The peptide mixture was analysed by a homemade 30 cm-long pulled-tip analytical column (75 μm ID packed with ReproSil-Pur C18-AQ 1.9 μm resin, Dr. Maisch). The column was then placed in-line with an Easy-nLC 1200 nano HPLC (Thermo Fisher Scientific) for mass-spectrometry analysis. The analytical column temperature was set at 55 °C during the experiments. The mobile phase and elution gradient used for peptide separation were as follows: 0.1% formic acid in water as buffer A and 0.1% formic acid in 80% acetonitrile as buffer B, 0–1 min, 6–10% B; 1–96 min, 10−36% B; 96–107 min, 36–60% B; 107–108 min, 60–100% B; 108–120 min, 100% B. The flow rate was set as 300 nl min−1.
Data-dependent tandem mass spectrometry (MS/MS) analysis was performed with a Q Exactive Orbitrap mass spectrometer (Thermo Fisher Scientific). Peptides eluted from the LC column were directly electrosprayed into the mass spectrometer with the application of a distal 2.2-kV spray voltage. A cycle of one full-scan MS spectrum (m/z 300–1,800) was acquired, followed by top 20 MS/MS events sequentially generated on the first to the twentieth most intense ions selected from the full MS spectrum at a 28% normalized collision energy. Full-scan resolution was set to 70,000 with an automated gain control (AGC) target of 3e6. MS/MS scan resolution was set to 17,500 with an isolation window of 1.8 m/z and an AGC target of 1 × 10−5. The number of micro scans was one for both MS and MS/MS scans, and the maximum ion injection time was 50 ms and 100 ms, respectively. The dynamic exclusion settings used were as follows: charge exclusion, 1, 2, 8 and >8; exclude isotopes, on; and exclusion duration, 5, 10 or 15 s. MS scan functions and LC solvent gradients were controlled by the Xcalibur data system (Thermo Fisher Scientific). Peptides containing the isopeptide bonds were identified using pLink2 software (pFind Team) as described previously37,38. Search parameters in pLink were: enzyme: trypsin; missed cleavages: 3; precursor and fragment tolerance: 20 ppm. Carbamidomethylation of cysteine was set as a fixed modification, and oxidation of methionine was set as a variable modification. The results were filtered by applying a 5% false-discovery-rate cut-off at the spectral level.
END-seq
Sequencing reactions were performed in a 35 μl volume of cleavage buffer (50 mM HEPES, pH 7.5, 30 mM KCl, 2 mM DTT and 1 mM MgCl2) containing 5 nM plasmid substrate with 400 nM wild-type or Y138F protein complex. The reactions were incubated for 60 min at 37 °C, then terminated by adding 0.5 μl of 500 mM EDTA and 0.8 μl of proteinase K (Roche, 20 mg ml−1), followed by an incubation for 30 min at 55 °C. NaCl was subsequently added to a final concentration of 150 mM. END-seq was performed as previously described, with modifications39. In brief, 12.5-day-old mouse testicular cells were washed twice in ice-cold PBS and resuspended in ice-cold cell suspension buffer. After equilibration at room temperature for 5 min, the resuspended testicular cells were combined with the cleavage reaction and embedded in 0.75% agarose (final concentration), solidifying at 4 °C for 15 min. The embedded cells were then lysed and digested with proteinase K (50 °C for 1 h, then 37 °C for 7 h). After washes with TE buffer (Tris-EDTA, pH 8.0), the plugs were treated with RNase at 37 °C for 1 h, followed by digestion with exonuclease VII (NEB) at 37 °C for 1 h and exonuclease T (NEB) at 24 °C for 45 min to blunt the DNA ends. After blunting, A-tails were added to the free 3′-OH ends, which were then ligated with END-seq hairpin adaptor 1. The agarose plugs were subsequently melted and dialysed, and the released DNA was sonicated to achieve DNA fragments of about 170 bp in length. These DNA fragments were collected and purified, then dissolved in 70 μl TE buffer. Biotinylated adaptor 1-DNA fragments were further isolated using MyOne streptavidin beads. The second end was repaired and A-tails were added, followed by ligation with END-seq hairpin adaptor 2. Both hairpin ends were digested with USER (NEB), and the resulting DNA fragments were amplified by PCR. The libraries were then sequenced using the Illumina HiSeq 2000 platform.
END-seq data analysis
Read1 reads were aligned to the mouse (GRCm38p2/mm10 plus plasmid) genome using Bowtie (v.1.2.1.1)40, allowing three mismatches and retaining only the best strata for reads with multiple alignments (-n 3 -k 1 -l 50). The ‘view’ and ‘sort’ functions in SAMtools (v.1.9) were used to convert and sort the mapping output, generating sorted BAM files41. BAM files were further converted to BED files using the bedtools (v.2.25.0) ‘bamtobed’ command42. To normalize read density (reads per million), the ‘genomecov’ function in bedtools (v.2.25.0) was applied, followed by conversion to a .bw file. Break positions were identified using the 5′ end of the reads.
Histological and immunohistochemical analyses
Testes from 19-dpp mice or ovaries from 12-dpp mice were fixed in either Bouin’s solution at room temperature or 4% paraformaldehyde (PFA) at 4 °C overnight, followed by embedding in paraffin and sectioning at a thickness of 5 μm. The sections fixed in Bouin’s solution were deparaffinized, rehydrated and stained with haematoxylin and eosin (H&E) for histological analysis. Each mouse was randomly selected on the basis of genotype, and the genotype was unknown during the analysis of fluorescent signals to ensure blinding. For immunofluorescence analysis, sections fixed in PFA were subjected to antigen retrieval by boiling in antigen retrieval buffer (10 mM sodium citrate, pH 6.0) for 18 min in a microwave oven, followed by cooling on ice for 30 min. Sections were then washed in PBS containing 0.1% Triton X-100 (PBST) and incubated in blocking buffer (10% donkey serum, 1% BSA in PBST) for 60 min at room temperature. Then the sections were incubated overnight at room temperature with rabbit anti-MVH (1:200, Abcam) in a blocking buffer. After washing in PBST, the slides were incubated with Alexa Fluor 594-conjugated donkey anti-mouse secondary antibody (1:500, Jackson ImmunoResearch) for 60 min at room temperature. After further washes in PBST, sections were mounted in Prolong Gold Antifade medium with DAPI (Molecular Probes) and imaged using a fluorescence microscope (Zeiss, Axio Scope A1) by Zen capture software (Zeiss, v.2.3 blue edition).
Spermatocyte chromosome spreading and immunofluorescence staining
Testes were collected from 19-dpp male mice for spermatocyte chromosome spreading43. Each mouse was randomly selected on the basis of genotype, and the genotype was unknown during the analysis of fluorescent signals to ensure blinding. In brief, seminiferous tubules were treated with hypotonic buffer (30 mM Tris, 5 mM EDTA, 50 mM sucrose, 17 mM trisodium citrate dihydrate and 0.5 mM dithiothreitol, pH 8.2) for 10–20 min, followed by gentle smashing in 100 mM sucrose buffer (pH 8.2). The resulting suspension was gently spread onto slides pre-coated with fixative buffer (1% PFA, 0.15% Triton X-100, pH 9.2) and air-dried in a humidity chamber at room temperature. After washing in PBST, the slides were incubated in blocking buffer (10% donkey serum, 1% BSA in PBST) for 60 min at room temperature. Then, the slides were incubated overnight at room temperature using the following primary antibodies: mouse anti-γH2AX (1:500, Millipore), mouse anti-SYCP3 (1:200, Santa Cruz), rabbit anti-SYCP3 (1:200, Abcam), rabbit anti-SYCP1 (1:200, Abcam) and rabbit anti-RPA2 (1:200; a gift from M. Luo). After washing in PBST, the slides were incubated for 60 min at room temperature with the following secondary antibodies: Alexa Fluor 488-conjugated donkey anti-mouse IgG (1:500, Jackson ImmunoResearch), Alexa Fluor 594-conjugated donkey anti-mouse IgG (1:500, Jackson ImmunoResearch), Alexa Fluor 488-conjugated donkey anti-rabbit IgG (1:500, Jackson ImmunoResearch) and Alexa Fluor 488-conjugated donkey anti-rabbit IgG (1:500, Jackson ImmunoResearch). After incubation with the corresponding secondary antibodies and washing in PBST, slides were mounted in Prolong Gold Antifade medium with DAPI (Molecular Probes) and imaged with a fluorescence microscope (Zeiss, Axio Scope A1) by Zen capture software (Zeiss, v.2.3 blue edition). For the quantification of γH2AX intensity, matched-exposure images were taken and signal intensities were measured by ImageJ.
Structural prediction using AlphaFold 3
AlphaFold 3 was used to predict the structures of the SPO11–TOP6BL heterodimer, the SPO11–TOP6BL tetramer and the SPO11–TOP6BL tetramer in complex with DNA44. The predicted three-dimensional structures were visualized using PyMOL software for detailed structural analysis44.
Statistics and reproducibility
A Mann–Whitney U test was used to evaluate the statistical significance of RPA foci or γH2AX intensity per cell between controls and mutants. P values were calculated using GraphPad Prism v.10. Time courses of linear DNA formation were fitted to an allosteric sigmoidal model in Prism v.10. The statistical significance of comparisons between wild-type and mutant SPO11 was assessed using a two-way ANOVA. For EMSA, binding curves were fitted individually with GraphPad Prism v.10 software, using a dose response that incorporated a Hill slope. Data were normalized to reflect the percentage of bound oligonucleotides and are presented as the mean ± s.d. of the interpolated Kd from three independent experiments. Each conclusion in the manuscript was based on results that were reproduced in at least three independent experiments and at least three mice of each genotype.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.