Human samples
Human intestinal tissue samples and annotated data were obtained, and experimental procedures performed, within the framework of the non-profit foundation HTCR (Munich, Germany), including informed patient consent. The framework of the HTCR Foundation has been approved by the ethics commission of the Faculty of Medicine in the Ludwig Maximilian University (no. 025-12) and the Bavarian State Medical Association (no. 11142). Twenty-seven consenting patients underwent visceral surgery with partial resection of intestine (16âmales, 11âfemales, age range 40â89âyears; precise intestinal regions are listed in Supplementary Table 4) for various oncologic indications, including pancreaticojejunostomy due to pancreatic adenocarcinoma (example no. ICD10, codes C18.x, C24.x and C25.x). We used micro- and/or macroscopically tumour-free regions of resectates for further preparation.
Isolation of TRM cells and donor-matched crypts for organoid generation
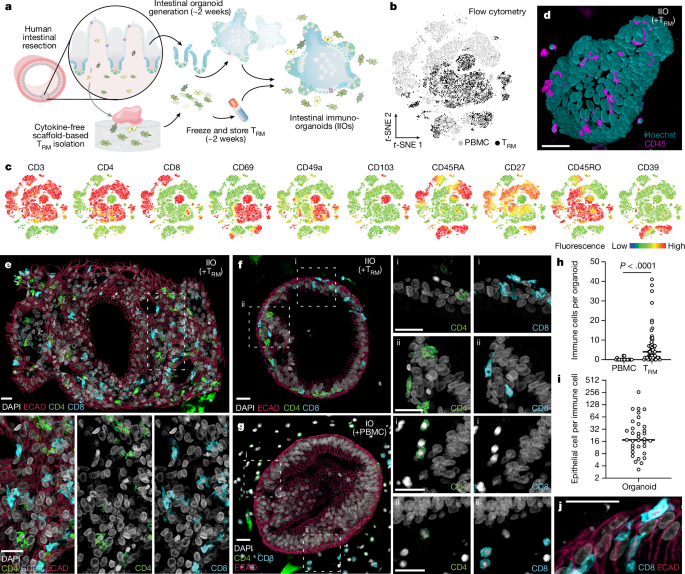
Intestinal tissue was first processed by removal of the underlying muscularis, serosa and fat from the basal side of the tissue by pinching with forceps and trimming away with scissors. The remaining mucosal tissue was washed with PBS (supplemented with penicillin (1,500âUâmlâ1), streptomycin (1,500âμgâmlâ1) and gentamicin (500âμgâmlâ1)) multiple times before using a scalpel to remove excess mucus from the luminal side and blood vessels from the basal side. Trimmed, cleaned tissue was then cut into square explants (approximately 5âÃâ5âmm3) with a scalpel. Lymphoid follicles such as Peyerâs patches were not separated from lamina propria tissue. Two methodologies were used to isolate intestinal TRM cells. For the scaffold-based egression protocol (adapted from ref. 20), each explant was loaded onto a 9âÃâ9âÃâ1.5âmm3 tantalum-coated, carbon-based scaffold (Ultramet) and cultured in 24-well plates containing 1âml of cytokine-free cell culture medium (RPMIâ1640, 10% fetal calf serum (FCS), penicillin (1,500âUâmlâ1), streptomycin (1,500âμgâmlâ1), gentamicin (500âμgâmlâ1) and amphotericin (12.5âμgâmlâ1)). Twenty-four hours later, scaffolds and tissue were removed and egressed cells were harvested from the bottom of the culture well via pipetting. For the enzymatic digestion protocol, explants were digested using the Human Tumour Dissociation Kit with the gentleMACS Octo Dissociator program 37C_h_TDK_1 (Miltenyi Biotec), as per the manufacturerâs instructions, reducing EnzymeâR content by 80% to minimize surface antigen cleavage. Cells were then counted, phenotyped by flow cytometry and frozen before use in downstream applications. For isolation of donor-matched crypts for organoid generation, a lamelle was scraped over a 3âcm2 piece of trimmed tissue to remove intestinal protrusions/villi. Tissue was then incubated in ice-cold PBSâ+â10âmM EDTA with vigorous shaking for 30âmin to break down epithelial cell junctions. Tissue was retrieved and a lamelle used to scrape off crypts into DMEM-F12 1% bovine serum albumin (BSA) collection buffer. Crypts were centrifuged, resuspended in Matrigel Matrix GFR (Corning) and cultured in IntestiCult Organoid Growth Medium with 10âμM Y-27632 (STEMCELL Technologies). Absolute authentication was confirmed by expression of cell lineage-defining markers and transcriptomic analysis. All lines used in the studies were verified as negative for mycoplasma before further experimentation.
Preparation and culture of IIOs, including treatment
In-passage organoids, approximately 2âweeks to 1âmonth following initial isolation, were cultured for 4âdays post split in IntestiCult Organoid Growth Medium and then switched to IntestiCult Organoid Differentiation Medium (STEMCELL Technologies) for 72âh to promote epithelial stem cell differentiation. On the day of co-culture set-up, medium was aspirated from the well and organoids were washed with PBS and treated with ice-cold Cell Recovery Solution (Corning) for 40âmin. The solution was collected and centrifuged to harvest the liberated organoids. Donor-matched TRM cells or PBMCs were thawed, combined with the liberated organoids and resuspended in either Matrigel Matrix GFR (Corning) or, if co-cultures were to be formalin fixed, a 50:50 mixture of Matrigel Matrix GF and 4âmgâmlâ1 Cultrex Rat CollagenâI (R&D Systems). For time-lapse live-imaging experiments, immune cells were labelled with CellTrace Far Red or CFSE (ThermoFisher) before mixing with organoids. Organoids were used at a concentration double their standard passaging density, whereby a 20âµl dome was harvested and resuspended in 10âµl of matrix, whereas immune cells were used at a density of 15,000âmmâ3 of resuspension volume. A 50:50 ratio of RPMIâ1640 10% FCS and IntestiCult Organoid Differentiation Medium was used for culture. For assessment of TCB-based cytotoxicity, an EpCAM-CD3 bispecific antibody, or its associated non-targeting control (which contains a CD3 binder on one arm and a non-specific DP47 arm), was supplemented into the culture medium following co-culture set-up, or 10âdays following co-culture set-up in the case of experiments with longer-term IIO cultures, typically at a concentration of 5ângâmlâ1. When used, blocking antibodies to TNF (adalimumab, InvivoGen) were added at a concentration of 1âµgâmlâ1. To investigate the role of ROCK signalling on TRM cell activation, 10âµM Y-27632 was added daily for the duration of the co-culture. For longer-term co-cultures (up to 1âmonth), a 50:50 ratio of RPMIâ1640 10% FCS and IntestiCult Organoid Growth Medium, supplemented with IL-2 (10âIUâmlâ1) (Roche) and IL-15 (2ângâmlâ1; BioLegend) was used. Medium was changed three times per week and the culture split once per week. Cultures were supplemented with 10âµM Y-27632 (STEMCELL Technologies) following splitting. For investigation of TRM cell-generated inflammation to study recruitment of circulating Tâcells into the epithelium, CellTracker CMFDA Green (ThermoFisher) was used to stain PBMCs at a final working concentration of 1âµM. For single TRM cellâorganoid and PBMCâorganoid co-cultures, the density of immune cells previously described was used. However, for TRM cellâPBMCâorganoid co-culture, both TRM cell and PBMC compartments were seeded at 15,000 cellsâmmâ3 of resuspension volume.
Flow cytometry analysis of immune-organoids
Triplicate wells of immuno-organoid co-cultures from each condition were harvested 5, 24 and 48âh post treatment. At 4âh preceding culture harvest at each time point, wells were treated with Protein Transport Inhibitor Cocktail (ThermoFisher) to facilitate intracellular accumulation of cytokine protein. Co-cultures were washed with PBS and then digested to single cells using Accutase solution (STEMCELL Technologies) at room temperature for approximately 30âmin. Cell suspensions were passed through a 70âμm strainer and stained for surface proteins (Supplementary Table 3). Cells were then fixed and permeabilized using the Foxp3 Transcription Factor Staining Buffer Set (ThermoFisher), and subsequently stained for intracellular and intranuclear proteins. Stained cell suspensions were acquired on a BD Fortessa X-20 using BD FACSDiva Software v.9.7 and analysed using FlowJo v.10. Example gating strategies are shown within the source data of the relevant figures in Supplementary Information. To facilitate visual representation across one plot, samples from different time points and treatments were concatenated and separated along the yâaxis.
Luminex supernatant analysis of immune-organoids
Following sampling, supernatants were immediately stored at â80â°C until measurement. For measurement of cytokines (Gzmb, IFNγ, IL-2, IL-4, IL-6, IL-10, TNF, MCP-1, IP-10 and GM-CSF) the customized Invitrogen ProcartaPlex multiplex immunoassay (reference no. PPX-10-MXFVMZC) was applied and used according to the manufacturerâs instructions. In short, capture beads were added to a 96-well flat-bottom plate, washed with an automatic plate washer (405TS microplate washer, Bioteck) and incubated with either the diluted supernatants or provided standards for 2âh at room temperature. Next the beads were washed and detection antibodies added to the plate for 30âmin at room temperature, followed by a further wash step. The beads were then incubated with Streptavidin-PE for a further 30âmin at room temperature before final washes. Lastly the beads were resuspended in the acquisition buffer and the plate was read on a Bio-Plex-200 instrument (Bio-Rad) using the corresponding Bio-Plex Manager Software v.6.2.
Isolation of Tâcell subsets for organoid co-culture
For organoid co-culture with specific subsets of immune cells, magnetic-activated cell separation MS columns and MicroBeads (Miltenyi Biotec) were used for subset enrichment following the manufacturerâs instructions. To enrich memory Tâcells from PBMCs, CD45RO MicroBeads were used (Miltenyi Biotec). To enrich for CD103+/â cells from TRMÂ cells, cells were first labelled with biotin anti-human CD103 antibody (BioLegend) and subsequently purified using anti-biotin MicroBeads (Miltenyi Biotec). Purity of the enriched cells was validated by flow cytometry.
Caspase 3/7-based epithelial cell cytotoxicity assay
Intestinal immuno-organoid cultures were prepared in 4âµl of Matrigel Matrix GF per well of a PhenoPlate 96-well microplate (Revvity), with cell ratios and media as described above. Apoptosis was assessed using the CellEvent Caspaseâ3/7 Detection Reagent (Invitrogen), either during TCB treatment or at specific intervals. CellEvent Caspaseâ3/7 Detection Reagent was added to culture medium at 1:1,000. Samples were imaged in confocal mode at Ã5âmagnification (air objective) with an Operetta CLS (Perkin Elmer) covering approximately a 450âµm z-stack, starting at â150âµm. Distance between z-stacks was set to the minimum of 27âµm for the Ã5âobjective (autofocus, two-peak; binning, 2). Channels selected were bright-field and predefined AlexaFluorâ488. Per well, five fields were acquired, covering nearly the entire surface of the 96-well PhenoPlate plate; CO2 was set to 5% and temperature to 37â°C. Caspaseâ3/7 fluorescence signal intensity was quantified using Opera Harmony software v.4.9 (PerkinElmer). Briefly, segmentation of organoids was done using âFind Texture Regionsâ based on the bright-field signal only, followed by âSelect Regionâ and âFind Image Regionâ to segment single organoids as objects. Next, âCalculate Morphology Parametersâ was performed to select objects above 10,000âµm2 with âSelect Populationâ. Next, Caspaseâ3/7 fluorescence signal per individual organoid was determined using âCalculate Intensity Propertiesâ of the AFâ488 channel within these objects.
Time-lapse imaging of IIOs
Before IIO or organoidâ+âPBMC-derived Tâcell co-culture, matched TRM cells and PBMCs were thawed and apoptotic cells removed using the Dead Cell Removal Kit (Miltenyi Biotec). Tâcells were isolated from PBMCs using the EasySep Human TâCell Isolation Kit (no. 17951, STEMCELL Technologies) and the EasySep Magnet (no. 18000, STEMCELL Technologies), following the manufacturerâs instructions. Immune cells were labelled with CellTrace Far Red (no. C34564, ThermoFisher) and IIO or PBMCâ+âorganoid co-cultures were prepared as described in âPreparation and culture of intestinal immune-organoids, including treatmentâ. Time-lapse live imaging was performed 16âh post co-culture set-up with a Leica STELLARISâ8 confocal microscope using a water-immersion objective (HC FLUOTARâL VISIR Ã25/0.95 numerical aperture WATER) and 0.85âzoom. Images were obtained in bidirectional mode with 1,024âÃâ1,024 pixels at 600âHz. Images were acquired every 38âs with 84âµm z-stacks (z-steps, 4âµm). Samples were imaged between 42 and 60âmin, where indicated. During imaging, samples were maintained in an incubation chamber (The Box, Life Imaging Services) at 37â°C and 5% CO2. Following acquisition, maximum-intensity projections were generated with Leica LasâX software and later exported as AVI files using ImageJ v.1.54i. Cell videos were analysed using CellProfiler v.4.2.5. The number of frames per condition was equalized for direct comparison. Briefly, cells were segmented at each frame with Multi-Otsu, morphological parameters were extracted and segmented cells were tracked over time using an overlap of two pixels. The output per cell was analysed using KNIME v.5.2.4. The means and standard deviation of morphological features per track over time were calculated. The standard deviation of morphological features over time is a measure of the dynamism of cells because they use amoeboid motility. In addition, track lifetime and track total distance were calculated. Tracks with fewer than 20âframes were discarded.
Monocyte-derived dendritic cell generation for ICI experiments
Monocyte-derived dendritic cells were generated using the Mo-DC generation Toolbox (Miltenyi Biotec) following the manufacturerâs instructions. Briefly, monocytes were isolated from PBMCs by magnetic separation over magnetic-activated cell separation columns with CD14 MicroBeads. Monocytes were cultured in standard tissue culture flasks in the provided Mo-DC Differentiation Medium for 7âdays, with renewal of the medium every 2â3âdays. Maturation of dendritic cells was initiated at dayâ7 for 3âdays by the addition of 6,000âIUâmlâ1 human TNF (Miltenyi Biotec). One day before usage, 100ângâmlâ1 lipopolysaccharide (Sigma-Aldrich) was added to activate dendritic cells overnight. Cells were harvested by incubation in 2ÃâEDTA in PBS for 10âmin at 37â°C. Dendritic cells were counted and plated at 40,000 per well of an ultralow-attachment, round-bottom, 96-well plate (Corning). TRM cells and PBMCs from the same donors were thawed, counted and combined with dendritic cells at 120,000âcells per well. Either immune checkpoint inhibitors (nivolumab and ipilimumab, both Bristol-Myers Squibb) or isotype controls were added at 20âµgâmlâ1, in RPMIâ1640 with 10% FCS. Dendritic/Tâcell co-cultures with or without ICIs were cultured for 4âdays before combining immune cells with matched organoids. Organoids were harvested as described in the preparation and culture of IIO before being mixed with the corresponding cells from the mixed lymphocyte reaction. IIO cultures were plated in 5âµl of Matrigel Matrix GFR domes and maintained in 50:50 RPMIâ1640 with 10% FCS (ThermoFisher) and IntestiCult Organoid Growth Medium (STEMCELL Technologies) for 48âh, with or without ICIs. Cell death was monitored by live imaging with Caspaseâ3/7, and LDH in the supernatant was measured every 24âh.
LDH assay
Supernatants of the triculture were used directly following sampling for measurement. A cytotoxicity detection kit (Roche) was used according to the manufacturerâs instructions. Briefly, the standard curve was prepared and supernatants were diluted in PBS and incubated with the reaction mix for 30âmin at room temperature in the dark. Following incubation the plate was read using a PerkinElmer Envision 2104âMultilable reader with absorbance at 490ânm.
Recombinant TNF treatment of organoid cultures
Four days following passage, organoids were harvested with cell recovery solution (Corning) and seeded onto a PhenoPlate 96-well microplate (Revvity) in 5âµl of Matrigel Matrix GFR droplets. Organoids were treated for 72âh with human recombinant TNF (Miltenyi Biotec; highest dose, 156ângâmlâ1, seven-dose titration, fivefold dilution). TNF-induced apoptosis was monitored by live imaging every 2âh on an Operetta using the cell event Caspaseâ3/7 detection reagent as described above.
FFPE embedding of co-cultures
To formalin-fixed, paraffin-embedded (FFPE) co-cultures the samples were seeded in a 50% (v/v) MatrigelâCollagenâI matrix. Wells were washed once with 1ÃâDPBS before fixation with 4% paraformaldehyde (PFA) in the 24-well Clear TC-treated plate. Following 30âmin of fixation at room temperature, the wells were washed three more times before complete aspiration of 1ÃâDPBS; 400âµl of preliquefied HistoGel (ThermoScientific) was then dispensed into 24-well Clear TC-treated plates. Following polymerization of HistoGel (10âmin at 4â°C), the organoidâHistoGel âplateletâ was carefully lifted out of the 24-well Clear TC-treated plate using a thin metallic spatula. Samples were then distributed into biopsy cassettes and dehydrated overnight using a Vacuum filter processor (Sakura, TissueTek VIP5). The following day, samples were embedded in liquid paraffin.
Microtome sectioning
FFPE blocks were, in general, sectioned at a thickness of 5âµm and transferred on Superfrost Plus Adhesion microscope slides (Epredia). Where indicated, thickness differs. Slides were incubated in a slide oven overnight at 37â°C.
FFPE-based mIF
Multiplex immunofluorescence (mIF) staining of FFPE slides was performed using a Ventana Discovery Ultra automated tissue stainer (Roche Tissue Diagnostics). Slides were first baked at 60â°C for 8âmin and subsequently further heated to 69â°C for 8âmin for subsequent deparaffinization. This cycle was repeated twice. Heat-induced antigen retrieval was performed with Tris-EDTA buffer pHâ7.8 (Ventana) at 92â°C for 32âmin. After each blocking step with Discovery Inhibitor (Ventana) for 16âmin, the Discovery Inhibitor was neutralized. Primary antibodies were diluted in Discovery Ab diluent (Ventana). Primaries were detected using appropriate anti-species secondary antibodies conjugated to horseradish peroxidase (HRP, OmniMap Ventana; Supplementary Table 3). Subsequently, the relevant Opal dye (Akoya Biosciences) was applied. Following every application of a primary, respective secondary antibody and Opal dye, an antibody neutralization and HRP-denaturation step was applied to remove residual antibodies and HRP before starting the staining cycle again with the Discovery Inhibitor blocking step. Lastly, samples were counterstained with DAPI (Roche).
mIF staining using Opal dyes from Akoya was digitized with multispectral imaging by Vectra Polari (PerkinElmer) using MOTiF technology at Ã20âmagnification for all seven colours (Opal 480, Opal 520, Opal 570, Opal 620, Opal 690, Opal 780 and DAPI). Slides were scanned in a batch manner to ensure identical imaging settings and cross-comparability for subsequent image analysis with HALO AI. Next, unmixing of channels and tiling of images was performed with PhenoChart (v.1.0.12) and inForm (v.2.4). Tiles were fused in HALO (Indica labs, v.3.2.1851.328).
High-resolution mIF was obtained using a STELLARISâ8 microscope (Leica) with a Ã40/1.1 numerical aperture water-immersion objective (HC FLUOTARâL VISIR Ã25/0.95 numerical aperture WATER) and 1.0âzoom. A white-light laser (440â790ânm) facilitated imaging of all Opal dyes mentioned above, and channels were acquired sequentially to reduce cross-talk. Images were obtained in bidirectional mode with 2,048âÃâ2,048âpixels (pixel size, 273.8âÃâ273.83ânm2) at 600âHz. Where indicated, images were acquired with z-stacks of 10â15âµm (z-steps, 1âµm), three-dimensionally reconstructed and shown in âMaximumâ mode using Leica Application SuiteâX software (Leica).
Image analysis by FFPE-based mIF
Image analysis of mIF images was performed with HALO AI (Indica Labs, v.3.2.1851.328). Briefly, single organoids were automatically detected using a deep learning algorithm trained to distinguish matrix and organoids (iterations, 5,000; cross-entropy, 0.32; DenseNet AI V2 Plugin). Following rapid validation, organoids were annotated as individual regions of interest, objects. Only objects over 7,500âµm2 were considered positive.
The HighPlex FL v.4.2.14 module was used to perform nuclear segmentation based on DAPI+ cells (assisted by HALOâs integrated AI-default ânuclear segmentation typeâ) and specific marker identification. For quantification, DAPI+ nuclei and markers for each distinct cell type of interest were merged (taking membranous and nuclear signals into account). Either secondary-only negative controls on the tissue of origin or organoid samples embedded within the same block served, and were then exposed to the full antibody-staining panel as a negative control sample to set the threshold for prevention of biased adjustments. The HighPlex FL analysis module was deployed on previously generated regions of interest of the organoids using integration of the classifier in the module. The number of Tâcells integrated/infiltrated into the organoid was normalized according to either the number of epithelial cells (non-TCB-treated organoids) or organoid area (EpCAM-targeted organoids). Surface markers of distinct Tâcells were used to phenotype these accordingly. Data was exported from HALO and analysed in GraphPad prism.
Single-cell dissociation of IIOs
Intestinal immuno-organoids were dissociated as described previously62. In short, organoids were dislodged, mechanically dismantled and transferred to 1% BSA-coated tubes. Organoid fragments were centrifuged at 400g for 4âmin at room temperature. The supernatant was removed and enzymes of the neural tissue dissociation kit (Miltenyi Biotec) were mixed in HBSS/1%BSA buffer. Organoid fragments were then dissociated to single cells for a total of 30âmin with thorough pipetting every 7âmin. Next, cells were filtered through a 40âμm filter, with single cells centrifuged at 450g for 4âmin at room temperature and subsequently resuspended in DPBS 1% BSA. Single-cell libraries were prepared on the 10X Genomics platform using the Chromium Next GEM Single Cell 3â²âKit v.3.1.
scRNA-seq data preprocessing
CellRanger (v.6.0.2, 10X Genomics) was used to extract unique molecular identifiers, cell barcodes and genomic reads from the sequencing results of 10X Chromium experiments. Next, count matrices, including both protein coding and non-coding transcripts, were constructed aligning against the annotated human reference genome (GRCh38, v.3.0.0, 10X Genomics). For removal of potentially damaged or unhealthy cells and improvement in data quality, the following filtering steps were performed in addition to the built-in CellRanger filtering pipeline. Cells associated with over 50,000âtranscriptsâusually less than 1% of the total number of samplesâwere removed; and cells associated with a low number of unique transcriptsâfewer than 500âunique transcripts detected (1% of the total number of samples)âwere removed. Cells with over 20% of mitochondrial transcripts were removed. Transcripts mapping to ribosomal protein coding genes, as well as to mitochondrial genes, were removed, together with transcripts detected in fewer than ten samples.
Normalization with SCTransform
For normalization and variance stabilization of the molecular count data of each scRNA-seq experiment we used the modelling framework of SCTransform in Seurat v.3 (ref. 63). In brief, a model of technical noise in scRNA-seq data is computed using âgeneralized gamma poisson regressionâ64. The residuals for this model are normalized values that indicate divergence from the expected number of observed unique molecular identifiers (UMIs) for a gene in a cell, given that geneâs average expression in the population and cellular sequencing depth. In addition, a curated list of cell cycle-associated genes, available within Seurat, was used to estimate the contribution of cell cycle and remove this source of biological variation from each dataset to increase the signal deriving from more interesting processes. The residuals for the top 2,000âvariable genes were used directly as input for computation of the top 100âprincipal components by principal component analysis (PCA) dimensionality reduction through the RunPCA() function in Seurat. Corrected UMIs, which are converted from Pearson residuals and represent expected counts if all cells are sequenced at the same depth, were log transformed and used for visualization and differential expression analysis.
Both primary intestinal biopsy samples and primary multiorgan biopsy samples were processed as described above. However, these did not undergo any cell filtering because quality control steps had already been performed in the respective published studies.
Doublet removal with DoubletFinder
For each scRNA-seq experiment, DoubletFinder65 (v.2.3.0) was used to predict doublets in the sequencing data. In brief, this tool generates artificial doublets from existing scRNA-seq data by merging randomly selected cells, which are then preprocessed together with real data and jointly embedded on a PCA space that serves as a basis for finding each cellâs proportion of artificial k-nearest neighbours. For this step we restricted the dimension space to the top 50âprincipal components. Finally, the proportions of artificial k-nearest neighbours values were rank ordered according to the expected number of doublets, and optimal cut-off selected through receiving operating characteristic analysis across pNâpK parameter sweeps for each scRNA-seq dataset: pN describes the proportion of generated artificial doublets, with pK defining principal component neighbourhood size. To achieve maximal doublet prediction accuracy the mean variance-normalized bimodality coefficient was leveraged. This provides a ground-truth-agnostic metric that coincides with pK values that maximize area under the curve in the data. To overcome DoubletFinderâs limited sensitivity to homotypic doublets, we consider doublet number estimates based on Poisson statistics with homotypic doublet proportion adjustment assuming 1/50,000âdoublet formation rate of the 10X Chromium droplet microfluidic cell loading.
Ambient messenger RNA signal removal
Following doublet prediction and removal we analysed each scRNA-seq dataset to estimate the extent of ambient mRNA contamination in every single cell and correct it. We used the Râpackage Cellular Latent Dirichlet Allocation (CELDA)66 (v.1.16.1), which contains DecontX and is a method based on the Bayesian statistical framework used to computationally estimate and remove RNA contamination in individual cells without empty droplet information. We applied the DecontX() function in CELDA to the raw count matrices with default parameters. Subsequently, we removed all cells with contamination values above 0.5 and used the decontaminated count matrices resulting from DecontX() for downstream analysis.
Geometric sketching
Geometric sketching is a downsampling technique that helps explore and interpret scRNA-seq data more effectively by providing a concise and intuitive representation of the cellular landscape that preserves rare populations. We used the sketchData() function from CellChat67, with default parameters, to select one-third of the sequenced cells for each donor in the homeostatic samples described in Extended Data Fig. 2 (scRNA-seq samples for donor no. 1â3). This strategy was used to avoid variability in sequence efficiencies that influence the computation of lower-dimensional embeddings and heterogeneity analysis.
Data integration
Individual datasetsâfollowing preprocessing, doublet removal and ambient mRNA regressionâwere aggregated according to specific criteria (for example, tissue of origin, profiling time, culture condition) and underwent a joint normalization step with SCTransform to mitigate technical confounding factors, which also served as a means for selection of a meaningful set of the 2,000âmost variable global genes before data integration. Integration of different conditions (culture model, treatment and time points) was performed using the log-normalized, corrected UMI count data in two steps. First, the residuals for the top 2,000âglobal variable genes were used as input in computing the top 100âprincipal components through the RunPCA() function in Seurat. The 30âleading principal components and 50ânearest neighbours were then used to define the shared neighbourhood graph with the FindNeighbors() function in Seurat. Subsequently, datasets were clustered according to the shared neighbourhood graph using the Louvain algorithm68 through the Seurat function FindClusters(), with resolution 0.8. Finally, we used these high-resolution clusters to define a restricted, noise-reduced and cell state-specific set of genes (âDifferential expression analysisâ). In the second step of the integration process we compiled a list consisting of the ensemble of the 30âtop differentially expressed genes for each cluster and used it to focus and repeat PCA dimensionality reduction. The first 30âprincipal component vectors of the new PCA space served as the basis for obtaining a two-dimensional representation of the data through UMAP69 implemented in RunUMAP() with the 50ânearest neighbours. We then computed a shared neighbourhood graph on the UMAP lower-dimensional space and computed the final integrated clusters with resolution parameter 0.2.
Integration index
The integration index of Tâcells in the homeostatic samples described in Fig. 2 and Extended Data Fig. 2 was computed by identification of the 50âclosest neighbours of each Tâcell on the lower-dimensional space defined by the 30âleading CCA vectors. Subsequently, the proportion of PBMC-derived Tâcell neighbours was subtracted from the proportion of IIO-derived Tâcell neighbours of each individual cell and the resulting index was mapped to an interval of 0â1.
Differential expression analysis
Gene differential expression analysis between distinct cell populations in scRNA-seq data was assessed by performing Wilcoxon rank-sum tests and area under the receiving operating characteristic analysis, as implemented by the Presto (v.1.0.0) package in R. log-transformed, corrected UMIs were used as input for differential expression statistical tests, and genes were called differentially expressed if the associated adjusted Pâvalue (Bonferroni method) was lower than 0.05, area under the curve value above 0.6 and log fold change greater than 0.15. In addition, we also set thresholds on detection rates of differential expression genes. In particular, a given gene was assigned as overexpressed in the analysed group if it was detected in at least 30% of the samples of that group, whereas detection rate in background samples was at most 70% of the detection rate of the analysed group.
CD8+ T cell activation trajectory reconstruction
For reconstruction of the continuum of the CD8+ Tâcell activation trajectory in IIO models challenged with bispecific antibodies, we took advantage of diffusion pseudotime as implemented in the destiny package (v.3.14.0) in R. In brief, diffusion pseudotime uses random-walk-based distance, computed on the leading eigenvectors of a transition matrix, to order scRNA-seq data according to differentiation stages44,70. We used the DiffusionMap() function in the destiny package on the space identified by the 30âleading principal component vectors of the integrated PCA embedding of CD8+ clusters. Pseudotime values were then computed with the DTP() function in destiny on the diffusion map object using default parameters. Similarly global pseudotime, following TNF perturbation simulation, was based on a random-walk approach on the cell-state transition matrix.
Intercellular communication analysis
For investigation of ligandâreceptor-mediated cellâcell communication during immune cell activation in our IIO models we focused on the signals exchanged between TH1 cells, activated T-bet Bâcells and CD8+ CTLs. For this analysis we extracted genes labelled as either ligands or receptors from curated databases53 and required that these genes be differentially expressed between the three populations under investigation, which facilitated retrieval of directional information about signal exchange. To gain insights into functional cellâcell communication we used the NicheNet (v.02.01.2000) pipeline, which considers the influence of sender-cell ligands on receiver-cell gene expression53. NicheNetâs analysis pipeline provided us with a ranking of predicted ligands that most probably affect gene expression in activated T-bet Bâcells and CD8+ CTLs, highlighting the role of critical TH1-secreted factors in driving immune cell phenotypes within IIOs.
Functional enrichment analysis
To gain an understanding of the mechanisms underlying phenotypes in our data, differentially expressed genes were analysed for Gene Ontology biological process enrichment using one-sided hypergeometric testing. Pâvalues were adjusted for multiple testing hypotheses by the Bonferroni method, and only those enrichment results below a 5% significance level threshold were considered. For this analysis we considered only those biological processes consisting of sets with more than 10 but fewer than 300âmapped genes.
In silico perturbation analysis
To simulate dynamic shifts in cell identity resulting from ligand signalling cascade activation we used Nichenetâs prior model53. The first step involved the generation of simulated values by applying the gene regulatory network as a function and propagating the relative changes in gene expression following k-nearest-neighbour imputation of the gene expression data. This iterative (three times) signal propagation enabled us to calculate the broad, downstream effects of ligand perturbation, thereby estimating the global transcriptional shift. The estimation of cell-identity transition probability was accomplished by comparing this gene expression shift with that of local neighbours, utilizing a likelihood-based dynamical model. By doing so we could establish a measure of how cell identities transition in response to ligand perturbation. Finally, the transition probabilities were transformed into a weighted local average vector map, encoding the simulated directionality of cell-state transition for each cell. This workflow results from an adaptation and integration of CellOracle71 and scVelo72 in Python (v.3.7).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.