Mice, diets and treatments
All animal experiments were performed in accordance with UK Home Office licences (70/8891, PP0604995, 70/8646, 70/8468 and PP8854860) and in accordance with the UK Animal (Scientific Procedures) Act 1986 and EU direction 2010. They were subject to review by the animal welfare and ethical review board of the University of Glasgow and the University of Newcastle upon Tyne. To minimize pain, suffering and distress to the animals, we used single-use needles and non-adverse handling techniques. Mice were housed under controlled conditions (specific-pathogen free, 12 h–12 h light–dark cycle, 19–22 °C, 45–65% humidity) with access to food and water ad libitum. We added environmental enrichments, in the form of gnawing sticks, plastic tunnels and nesting material to all of the cages. Welfare of animals was defined by clinical symptoms, including visible masses, any degree of reduced mobility/distress, weight loss or evidence of haemorrhage; however no maximal tumour volume end points for intrahepatic tumours were mandated. No mouse exceeded the humane end points stipulated in the Home Office Licenses.
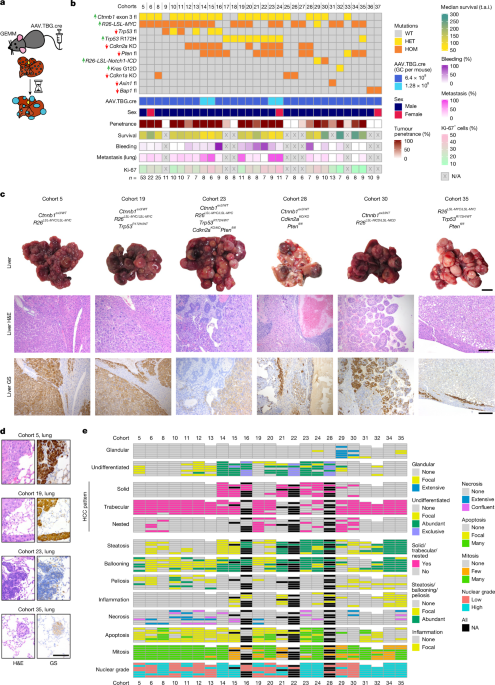
Unless otherwise specified, male mice on a mixed background were used. The following transgenic mice strains were used: Gt(Rosa)26Sortm14(CAG-tdTomato)Hze (R26LSL-Tom)44, Ctnnb1tm1Mmt (Ctnnb1ex3)45, Gt(Rosa)26Sortm1(MYC)Djmy (R26LSL-MYC)46, Trp53tm1Brn (Trp53fl)47, Trp53tm2Tyj (Trp53R172H)48, Cdkn2atm1.1Brn (Cdkn2aKO)49, Ptentm2Mak (Ptenfl)50, Gt(Rosa)26Sortm1(Notch1)Dam (R26LSL-NICD)51, Krastm4Tyj (KrasG12D)52, Cdkn1atm1Led (Cdkn1aKO)53, Axin1fl (ref. 54), Bap1tm2c(EUCOMM)Hmgu (ref. 55). Genotyping was performed by Transnetyx using ear notches taken for identification purposes at weaning (3 weeks of age). Mice were induced between 8 and 12 weeks of age, unless otherwise indicated, with AAV8.TBG.PI.eGFP.WPRE.bGH (AAV8-TBG-GFP) (Addgene, 105535-AAV8), AAV8.TBG.PI.cre.rBG (AAV8-TBG-cre) (Addgene, 107787-AAV8) or AAV8.TBG.PI.Null.bGH (AAV8-TBG-Null) (Addgene, 105536-AAV8). Virus was diluted in 100 µl PBS to the desired concentration and injected through the tail vein. Unless otherwise specified, mice received a dose of 6.4 × 108 GC per mouse.
For the GEMM + MWD model, 6-week-old mice were kept on a modified western diet (Envigo, TD.120528) plus sugar water (23.1 g l−1 fructose and 18.9 g l−1 glucose) in combination with repeated CCl4 injections (intraperitoneal (i.p.), 0.2 µl g−1 of body weight; vehicle, Cornoil) as previously described56 and were induced with AAV.TBG.cre at 10 weeks of age.
For the DEN/ALIOS model, C57BL/6 WT mice, were injected with a single dose of DEN (80 mg per kg by i.p. injection) at 14 days of age. Mice were fed ALIOS diet (Envigo, TD.110201) and sugar water (23.1 g l−1 fructose and 18.9 g l−1 glucose) from 60 days of age. Mice were collected at day 284.
For the MWD + CCl4 model, the mice were kept on a modified western diet (Envigo, TD.120528) plus sugar water (23.1 g l−1 fructose and 18.9 g l−1 glucose) in combination with repeated CCl4 injections (i.p., 0.2 µl g−1 body weight; vehicle, Cornoil) as previously described56.
For the streptozotocin (STZ) model, male and female C57BL/6J WT mice were injected with a single dose of STZ (200 µg in 0.1 M citrate buffer, pH 4.0) subcutaneously at 2 days of age. Mice were fed a high-fat diet (TestDiet 58R3, 1810835) from 30 days of age. All STZ–HFD-treated livers showed pale yellow colour at 6 weeks, mild swelling at 8 weeks, granular surface at 12 weeks and tumour protrusion at 20 weeks of age57. Mice were collected between 17 and 35 weeks of age.
For the orthotopic model, Hep-53.4 cells (female C57BL/6J hepatoma cell line; Cytion, LOT-L230232R) were injected intrahepatically into the left lobe of male C57BL/6J mice. The procedure was performed under isoflurane general anaesthesia. Analgesia was given to the mice for pain management. Mice were collected at 28 days after implantation or left to reach an approved humane end point.
For therapeutic intervention in the BM model, drugs were given at 90 days after induction or in other models determined by mean cohort survival relative to the BM model survival as indicated in the figures. The following drugs were used: sorafenib (LC Laboratories S8502, daily, oral gavage, 45 mg per kg; vehicle, 50% chremophor/50% ethanol, then, before dosing, 3 parts H2O added), lenvatinib (SelleckChem, S1164 (end-point studies); or Eisai (monotherapy timepoint studies); daily; oral gavage, 10 mg per kg, vehicle, 3 mM HCl), anti-PD1 (BioLegend, RMP1-14; twice per week; i.p., 200 µg; vehicle, PBS; control, IgG, BioLegend, RTK2758), cladribine (SelleckChem, S1199; daily; i.p., 20 mg per kg; vehicle, PBS), VEGFRi (AstraZeneca, AZD2171; daily; oral gavage, 3 mg per kg; vehicle, 0.5% (w/v) HPMC, 0.1% Tween-80, in H2O). To help with drug-induced weight loss, mice on cladribine treatment received irradiated peanuts and sunflower seeds as diet supplements. If mice reached 83% of their weight at treatment start, cladribine treatment was withheld until they gained weight to at least 90% of weight at treatment start. Mice who dropped below 80% of weight at treatment start were sampled according to licence limitations. Confounding factors (for example, litter mates, induction date) were taken into consideration when allocating mice into groups but mice were not randomized using a specific method. Mice who presented with a visible tumour before treatment start were excluded from the experiments according to a priori established criteria. Animal technicians dosing the mice were blinded to the genotype of the mice. The number of biological replicates was ≥3 mice per cohort for all experiments. Further details are provided in the figure legends and Supplementary Table 1.
Animal tissue collection
GEMMs were sampled at specific timepoints or at the end point. The end point was defined as the mouse having reached a liver weight/body weight ratio of >20% or having adverse side effects from the tumour, such as tumour haemorrhaging. Mice who died of tumour haemorrhaging were included in the survival analysis but not in any downstream analysis. Tumours were measured macroscopically using digital callipers, and visible tumours were counted. Images of whole livers were taken using a Canon PowerShot G9X camera with a ruler present in each picture. Tissue was either sampled in neutral buffered saline containing 10% formaldehyde or snap-frozen on dry ice.
Histology and immunohistochemistry
Liver, tumour and lung tissues were fixed using neutral buffered saline containing 10% formaldehyde, dehydrated and embedded in paraffin, and cut into 4-μm-thick sections. The sections were dewaxed and stained with H&E or Sirius Red using standard protocols. Additional sections were stained immunohistochemically using the primary antibodies listed in Supplementary Table 5. Primary antibodies were detected by HRP-labelled secondary antibodies and subsequently stained using a peroxidase DAB kit; either Agilent (K3468) or Leica (DS92563) DAB for tissue processed in autostainer or Vector Laboratories (SK-4100) with haematoxylin as a counterstain (immunohistochemistry) or by fluorescent-labelled secondary antibodies (Invitrogen) with DAPI used as counterstain (SouthernBiotech, 0100-20) (immunofluorescence).
Microscopy and quantitative analysis of immunohistochemistry
Images were obtained on the Zeiss Axiovert 200 microscope using the Zeiss Axiocam MRc camera. For image analysis, stained slides were scanned using the Leica Aperio AT2 slide scanner (Leica Microsystems) at ×20 magnification. Quantification of blinded, stained sections (GS, Ki-67, CC3, CD3, yH2AX, p53) was performed using the HALO image analysis software (v.3.1.1076.363, Indica Labs). Quantification of microscopy tumour area in BM mice was performed based on nodules, independent of GS status. Quantification of Ki-67+ was by percentage/cell number and CC3+ by percentage/tumour area.
Lungs were microscopically analysed for the presence of extrahepatic HCC spread by examining H&E and GS sections. Metastasis was scored in a binary manner as detected or not-detected but was not analysed in respect to individual metastasis burden per mouse.
Images for tissue comparison to HCCOs were taken on the Zeiss 710 confocal microscope.
Tumour genotyping
After extraction from whole tumour, DNA was suspended in Transnetyx assay buffer and was analysed by Transnetyx using probes (p53Flox EX, Bap1-2 EX, PTEN-EX, LSL-EX-1, Tg-MYC, Axin1-1 EX, Ctnnb1-16 EX) and was additionally purified and concentrated using the Monarch Genomic DNA purification kit (New England Biolabs, T3010L) according to the manufacturer’s instructions. Generation of amplicons indicating successful recombination of genetic loci was performed by PCR (Eppendorf Mastercycler x50a) using the OneTaq Quick-Load 2× Master Mix with Standard Buffer (M0486S); reactions were set up according to the manufacturer’s instructions, amplification conditions (Supplementary Table 4) and primer sequences—β-catenin exon 3 (Supplementary Table 4) and KRASG12D (Supplementary Table 4). The resulting PCR reactions were separated by electrophoresis on 1.5% agarose (Melford, A20090-500) gel, using the size marker Quick-Load Purple 1 kb Plus DNA Ladder (New England Biolabs, N0550S) and bands were visualized using SYBR Safe DNA gel stain (Invitrogen, S33102). The Gels were imaged on the Chemi-Doc Imaging System (Bio-Rad). Concordance between CTNNB1 recombination results between the two methods was 100%. Where possible, the samples used in histological comparison were also assessed genotypically. Where not possible, due to DNA contamination/low quality, tumours were replaced by other end-stage tumours from the same cohort to achieve n ≥ 6 per cohort (total n = 4 additional samples).
Quantitative analysis of fluorescence immunohistochemistry
Fluorescent tiled images were generated on the Opera Phenix High-Content Screening System (Perkin Elmer) at ×20 magnification. Fluorescence was detected using the same settings throughout. Consecutive, non-overlapping fields were analysed blindly using Columbus Image analysis software (v.2.8.0.138890, Perkin Elmer). Positivity gating thresholds were defined using negative controls. For representative images, processing adjustments were performed equally.
Multiplex immunofluorescence immunohistochemical staining
Mouse liver samples (thickness, 4 µm) were sectioned and placed onto TOMO hydrophilic adhesive microscope slides (Matsunami, 0808228600). After antibody validation, semi-automated multiplex immunofluorescence staining was performed on the Ventana Discovery Ultra platform (Roche Tissue Diagnostics, RUO Discovery Universal v.21.00.0019). Fluorescence detection was performed using an Opal fluorophore tyramide-based signal amplification system (Akoya Biosciences). All primary antibodies were optimized using a pH 9 antigen retrieval solution (CC1, Roche Tissue Diagnostics, 06414575001) at 95 °C for 32 min. A denature step was applied using pH 6 antigen retrieval solution (CC2, Roche Tissue Diagnostics, 05279798001) for 24 min between each Opal detection and primary antibody application.
The primary–secondary–opal fluorophore combinations (CD45–HRP–Opal480; CD8–HRP–Opal690; CD4–HRP–Opal620; GranzymeB–HRP–Opal650; GS–HRP–Opal520; MYC–HRP–Opal570) are described in Supplementary Table 5. ImmPRESS rat and Opal 780 were manually applied in their specific sequences, and the remaining reagents were fully automated on the Ventana DISCOVERY ULTRA platform (Roche Tissue Diagnostics). Three drops of nuclear DAPI counterstain (Roche Tissue Diagnostics, 05268826001, RTU (Ready to Use), 24 min) were applied to each sample for nuclear detection.
Multiplex immunofluorescence image acquisition and analysis
Whole-slide images were collected on the PhenoImager HT multispectral slide scanner (Akoya Biosciences, v.1.0) using a ×10 objective before the acquisition of each region of interest (ROI) using a ×20 objective. Each ROI was spectrally unmixed using InForm (Akoya Biosciences, v.2.6.0) using a project-specific spectral library created using single-channel dyes and an autofluorescence mouse liver control.
Visiopharm was used for all image analysis. For tissue segmentation, a bespoke, in-house-trained deep learning algorithm (v.2024.06.0.19093 ×64) was trained using the deep learning module with a U-Net backbone, to segment each lobe into tumour, stroma, non-tumour GS and background ROIs. Tumour regions that were smaller than 10,000 µm2 were classified as a ‘clone’ region. Tumour and clonal regions were then dilated to generate ‘peritumoural stroma’ and ‘periclonal’ regions, respectively. Necrotic regions were manually segmented. Tumour regions were eroded to create ‘tumour centre’ and ‘tumour periphery’ regions. T cells were classified with a ‘T cell’ label within these regions using the threshold module (v.2024.07.1.16745 ×64) to threshold CD4, CD8 and CD45 fluorescence channels using the original image features. Each image was verified by a pathologist to confirm regional and T cell label segmentation. Post-processing steps were included to change T cell labels into their respective regional labels, that is, a T cell found within the tumour ROI would be changed to ‘tumour T cell’ and so on. Output variables were then generated and exported for downstream data analysis. Area of entire lobes were generated, and areas for each ROI as well as regional mean pixel intensities of MYC, mean pixel intensities of each marker in each T cell label and x–y coordinates of each T cell label. The Phenoplex Guided workflow was used for T cell phenotyping, which generated a phenotype list that was exported for data analysis.
Duplex immunofluorescence immunohistochemical staining
For duplex immunofluorescence immunohistochemical staining (Extended Data Fig. 12b (bottom)), 4-µm-thick mouse HCCOs and liver lobe samples were sectioned and placed onto TOMO hydrophilic adhesive microscope slides (Matsunami, 0808228600). After antibody validation, fully automated multiplex immunofluorescence staining was performed on the Ventana DISCOVERY ULTRA platform (Roche Tissue Diagnostics, RUO Discovery Universal v.21.00.0019). Fluorescence detection was performed using an Opal fluorophore tyramide-based signal amplification system (Akoya Biosciences). All primary antibodies were optimized using a pH 9 antigen retrieval solution (CC1, Roche Tissue Diagnostics, 06414575001) at 95 °C for 32 min. A denature step was applied using pH 6 antigen retrieval solution (CC2, Roche Tissue Diagnostics, 05279798001) for 24 min between each Opal detection and primary antibody application. The primary–secondary–opal fluorophore combinations (MYC–HRP–Opal570, GS–HRP–Opal520) are described in Supplementary Table 5. One drop of nuclear DAPI counterstain (Akoya, 232121) was applied to each sample for nuclear detection.
Duplex immunofluorescence image acquisition and analysis
Whole-slide images were collected on the PhenoImager HT multispectral slide scanner (Akoya Biosciences, v.1.0) using a ×20 objective using Motif mode. Images were spectrally unmixed using Inform (Akoya, v.2.6.0) using an autofluorescence liver control slide to remove autofluorescence.
Tumour scoring
H&E-stained sections and tumours were additionally assessed by a consultant liver histopathologist and UK liver pathology External Quality Assessment scheme member (T.J.K.) working at a national liver transplant centre. All assessment was undertaken blind to all other data, including genotype and sampling times. An initial screen of the first available 135 cases was made to identify prominent histological features in lesional and non-lesional liver that could be semi-quantitatively assessed.
Accepting the inherent limitations of semi-quantitative subjective histological assessment but using a single observer to remove interobserver considerations, semi-quantitative/ordinal scoring systems were created for lesional and non-lesional features. Slides containing transections of whole lobes from each animal were assessed as a whole, giving an overall score or impression rather than scoring on an individual-lesion basis.
Non-lesional liver was scored for steatosis (none, focal, abundant) and lobular inflammation (none, focal, abundant). A minority of slides included insufficient non-lesional liver for assessment.
For lesional assessment, the presence of glandular tumour, that is, meriting designation as adenocarcinoma (none, focal, extensive) and undifferentiated carcinoma (none, focal, abundant, exclusive) was assessed first. All hepatocellular neoplastic lesions had the morphological and cytological appearances of malignancy, that is, HCC. In all cases in which there was HCC, the following features were assessed using the categories in parentheses: lesional pattern (any from nested, trabecular, solid), lesional steatosis (none, focal, abundant), lesional cell ballooning (none, focal, abundant), intralesional inflammation (none, focal, abundant), lesional necrosis (none, focal, confluent, extensive), lesional cell apoptosis (none, focal, many), intralesional peliosis (none, focal, abundant), lesional nuclear grade (low, minimal/low pleomorphism; high, highly pleomorphic).
Whole-tumour RNA-seq
Whole tumour and healthy tissue were snap-frozen and stored at −80 °C. To cover the breadth of our models, for each cohort, tissue from the shortest and longest surviving mouse as well as tissue from mice with survival closest to median cohort survival was chosen. Tissue was homogenized using the Precellys Evolution homogenizer and bulk RNA was isolated using a Trizol (Invitrogen) extraction protocol according to the manufacturer’s instructions. RNA quality and quantity was analysed on the Nanodrop 2000 (Thermo Fisher Scientific) and Agilent 2200 TapeStation (D1000 screentape) systems. Only samples with RIN > 7 were used for library preparation. Libraries were prepared using a Lexogen QuantSeq FWD Kit (disease positioning) or the Illumina TruSeq stranded mRNA kit (tumour heterogeneity). Library quality and quantity were assessed using the 2200 TapeStation (Agilent) and Qubit (Thermo Fisher Scientific) systems. The libraries for the disease positioning were sequenced by Novogene Europe. The libraries for the tumour heterogeneity were run on the Illumina NextSeq 500 system using the high-output 75 cycle kit (2 × 36 cycle paired-end reads).
Mapping of RNA-seq expression data
Quality checks and trimming on the raw RNA-seq data files were done using FastQC v.0.11.9 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/), FastP (v.0.20.1)58, MultiQC (v.1.9)59 and FastQ Screen (v. 0.14.0)60. RNA-seq single-end reads were mapped to the GRCm39.103 version of the Mus musculus genome and annotated61 using STAR (v.2.7.8a)62. Expression levels were determined by FeatureCounts from the Subread package (v.2.0.1)63.
Computational disease positioning based on human TCGA data
TCGA data were downloaded using the GenomicDataCommons R package (v.1.12.0; https://bioconductor.org/packages/GenomicDataCommons)64, TCGA ‘HTSeq–counts’ and corresponding clinical annotations. TCGA mutational data were downloaded using maftools (v.2.4.2)65. Both human and mouse RNA-seq counts were normalized using VST from the DESeq2 (v.1.28.1 and v.1.44.0) package66 and then centred within a sample. Genes were reduced to those with direct one-to-one gene mapping between human and mouse genomes established by Ensembl, as retrieved from the biomaRt (v.2.56.1) package67,68. Singular-value decomposition (SVD) of the human data was performed followed by matrix factorization of both the human and mouse data into a 100-rank human space. UMAP of the combined dataset was executed using R package uwot (v.0.1.11; https://CRAN.R-project.org/package=uwot). An adjacency matrix was constructed from a nearest-neighbours search (RANN package v.2.6.1, https://CRAN.R-project.org/package=RANN) of the human and mouse SVD objects for clustering analysis. R package igraph (v.1.2.11 and v.2.0.3)69 was used to construct a graph object and the community structure was determined using Louvain •clustering.
Single-sample gene set enrichment analysis (ssGSEA) analysis was performed using the R package corto (v.1.2.4)70 with the Hallmark gene set71,72 downloaded using msigdbr (v.7.4.1; https://CRAN.R-project.org/package=msigdbr). Hoshida27 and Chiang28 (also downloaded using msigdbr) subclass classification was determined by the highest enriched subclass. Tumour immune cell estimation was performed using ConsensusTME73.
Visualization of data by a combination of the ComplexHeatmap (v.2.4.3 and v.2.14.0)74, ggplot2 (v.3.3.6 and v.3.5.1)75, cowplot (v.1.1.1; https://CRAN.R-project.org/package=cowplot) and viridis76 packages.
Human H&E-stained tissue sections were obtained from the TCGA collection (https://portal.gdc.cancer.gov/).
Validation of HuMo clusters in an independent HCC cohort
HuMo clusters were validated with the bulk RNA-seq data of an independent cohort of 171 HCC samples from patients undergoing resection collected in the setting of the HCC Genomic Consortium25 (European Genome-Phenome Archive: EGAS00001005364). Fastq files were aligned using STAR62 (v.2.5.1b) to the hg19 reference genome with gencode annotation v19 and were quantified using featureCounts63 (v.1.5.2). Raw counts were preprocessed and cluster attribution was performed as described above with the TCGA and mouse data. In the analysis of the transcriptomic data, positivity for previously reported gene signatures was evaluated using the Nearest Template Prediction77 module from GenePattern (v.3.9)78. The ssGSEA projection71 was performed using previously reported gene signatures as well as the Hallmark gene set downloaded using msigdbr (v.7.4.1). The mutational profile of 144 HCC samples was obtained by whole-exome sequencing25. Clinicopathological data (such as vascular invasion, AFP levels (≥400 ng ml−1)) were originally reported previously25.
Differential expression analysis for intertumoural heterogeneity
Genes were restricted to those with significance in all comparisons (with significance defined as adjusted P < 0.05 and log2[FC] > 1). Data were scaled and visualized using the ComplexHeatmap74 package. Gene Ontology over-representation analysis was performed using the clusterProfiler79 package (v.3.16.1).
Human sample ethical approval
The use of consenting patients’ tissues surplus to diagnostic requirements for research purposes was approved by the Newcastle and North Tyneside Regional ethics committee, the Newcastle Academic Health Partners Bioresource (NAHPB) and the Newcastle upon Tyne NHS Foundation Trust Research and Development (R&D) department, in accordance with Health Research Authority guidelines. (10/H0906/41; NAHPB Project 48; REC 12/NE/0395; R&D 6579; Human Tissue Act licence 12534).
MRI
Magnetic resonance imaging (MRI) scans were performed on liver-tumour-bearing mice using the nanoScan imaging system (Mediso Medical Imaging Systems). The mice were anaesthetized and maintained under inhaled isoflurane anaesthesia (induction, 4–5% (v/v); maintenance, 1.5–2.0% (v/v)) in 95% oxygen during the entire imaging procedure. Whole-body T1-weighted gradient echo (GRE) 3D coronal/sagittal MRI sequences (echo time (TE), 3.8 ms; repetition time (TR), 20 ms; flip angle, 30 degrees; and slice thickness, 0.50 mm) were used to obtain MRI images. For quantification of scans, volumes of interest were manually drawn around the liver region on MRI scans by visual inspection using VivoQuant software (v.4.0, InviCRO). For each scan, separate volumes of interest were prepared to adjust for the position and angle of each mouse on the MRI scanner and their tumour size.
Mouse HCCO culture, drug screening and imaging
HCCOs were extracted and cultured as previously described31,80, with the exception that HCCOs from mice with activated β-catenin signalling were cultured in the absence of WNT and RSPO1. All mouse HCCO cultures were regularly tested for mycoplasma.
For the high-throughput screen cohort 5 (BM) HCCOs were dissociated with TrypLE and plated at a density of 1 × 103 cells in 10 μl BME in prewarmed 384-well plates (Greiner BioOne, 781091) 5 days before adding the drugs. On day 0, a panel of 147 FDA-approved oncology drugs (AOD IX, acquired June 2019, https://dtp.cancer.gov/organization/dscb/obtaining/available_plates.htm) was added at a final concentration of 10 µM. Staurosporin was used as an internal positive control; DMSO and untreated cells were used as an internal negative control. The medium was changed on day 4 and the compounds were freshly added. Incucyte NucLight Rapid Red (Sartorius, 4717) was added on day 6 and cells were imaged using the Opera Phenix High-Content Screening System (Perkin Elmer) on day 9. Volumes were determined using Icy BioImage software (v.2.0.0.0; https://icy.bioimageanalysis.org)81. The experiment was performed twice (using different passages from one HCCO line) in technical quadruplicates.
For the drug dose–response curve screen, HCCOs (1–2 lines per cohort) were dissociated with TrypLE and plated at a density of 1 × 103 cells in 10 μl Matrigel (Corning, 356231) in prewarmed 96-well plates (Greiner BioOne, 655098). The treatment schedule was the same as for the HTP screen, except the medium was changed and fresh drugs were added on days 3 and 7. Drugs and concentrations are shown in the figures. Drugs were purchased from Selleckchem, dissolved in DMSO to 10 mM, aliquoted and stored at −20 °C. Cell viability was measured on day 9 using CellTitre-Glo 3D reagent (Promega, G9682) according to the manufacturer’s instructions. Luminescence was measured on the Spark Microplate Reader (Tecan). The results were normalized to the vehicle. Curve fitting and IC50 calculation were performed using a nonlinear regression equation. All of the experiments were performed in duplicate and at least three times using different passages from one to two HCCO lines per cohort.
Images of HCCOs were taken on an Olympus CKX41 using the Qimaging Retiga Exi Fast 1394 camera.
For immunofluorescence analysis, HCCOs were washed with ice-cold PBS, fixed with 4% PFA and permeabilized with 0.2% Triton X-100. A list of the antibodies used is provided in Supplementary Table 5. Images were taken using the Zeiss 710 confocal microscope.
Tumour-derived mouse HCCOs (available from all GEMMs) will be shared on reasonable request.
Human HCCO culture and drug screening
Human HCCOs were derived from liver cancer needle biopsies or liver resections as described before32. The following human HCCO lines were used: D386-O and D953-O (CTNNB1 WT, TP53 WT, MYC WT); D455-O (CTNNB1 MUT, TP53 WT, MYC AMP); C948-O and C949-O (CTNNB1 MUT, TP53 WT, MYC AMP); C655-O (CTNNB1 WT, TP53 MUT, MYC ND); D045-O, D046-O, D803-O and R035-O (CTNNB1 WT, TP53 MUT, MYC WT); C798-O, C975-O, D324-O, D804-O and D876-O (CTNNB1 WT, TP53 MUT, MYC AMP); and D359-O (CTNNB1 MUT, TP53 MUT, MYC WT).
For expansion, the human HCCOs were seeded into reduced growth factor BME2 (R&D Systems, 3533-005-02) and cultured in expansion medium (EM): advanced DMEM/F-12 (Gibco, 12634010) supplemented with 1× B-27 (Gibco, 17504001), 1× N-2 (Gibco, 17502001), 10 mM nicotinamide (Sigma-Aldrich, N0636), 1.25 mM N-acetyl-l-cysteine (Sigma-Aldrich, A9165), 10 nM [Leu15]-gastrin (Sigma-Aldrich, G9145), 10 μM forskolin (Tocris, 1099), 5 μM A83-01 (Tocris, 2939), 50 ng ml−1 EGF (Peprotech, AF-100-15), 100 ng ml−1 FGF10 (Peprotech, 100-26), 25 ng ml−1 HGF (Peprotech, 100-39), 10% RSPO1-conditioned medium (v/v, homemade). HCCOs were passaged after dissociation with 0.25% trypsin-EDTA (Gibco). All human HCCOs were regularly tested for mycoplasma contamination using the MycoAlert mycoplasma detection kit (Lonza, LT07-118).
Drugs were purchased from ApexBio and Selleckchem, dissolved in DMSO to 10 mM, aliquoted and stored at −20 °C. For the screening, human HCCOs were dissociated with 0.25% trypsin-EDTA (Gibco) to single cells and 1 × 103 cells per well were plated in a 384-well plate (Greiner BioOne, 781986) on a layer of BME2 (R&D Systems, 3533-005-02) previously diluted with EM (50:50, v/v). Cells were cultured for 3 days without treatment to allow for organoid formation. At day 3, an eight-point half-log dilution series of each compound (ranging from 10 μM to 0.00316 μM) was added using a Tecan D300e. Cell viability was measured after 5 days of treatment using the CellTiter-Glo 3D reagent (Promega, G9682). Luminescence was measured on the Synergy H1 multi-mode reader (BioTek Instruments). Results were normalized to the vehicle (DMSO). The maximal DMSO concentration was 0.2%. Curve fitting was performed using Prism (GraphPad v.9 GraphPad Software) software and the nonlinear regression equation. Results are shown as mean ± s.e.m.
Fluorescent activated cell sorting
After mincing into small pieces, 100 mg of healthy liver or liver tumour was digested on the gentleMACS Octo dissociator with heaters using the mouse tumour dissociation Kit (Miltenyi Biotec, 130-096-730). Dissociated cells were resuspended in 0.5% BSA in PBS, filtered through a 70 μm strainer and centrifuged at 400 g for 5 min. Cells were then resuspended in 5 ml RBC lysis buffer (Thermo Fisher Scientific, 00-4300-54) and incubated for 5 min at room temperature and washed with 0.5% BSA in PBS before being resuspended in 0.5% BSA in PBS. Cell suspensions were added to 96-well V-bottom plates (maximum density, 0.5 × 106 cells per well). Cells were stimulated for 3 h with complete IMDM medium containing 8% FCS, 50 μM β-mercaptoethanol, 1× penicillin–streptomycin with 1× cell activation cocktail with brefeldin A (BioLegend, 423304) at 37 °C as previously described previously82. After stimulation, cells were centrifuged at 800g for 2 min. Cells were stained in Brilliant stain buffer (BD Biosciences, 566349) containing antibodies for surface antigens for 30 min at 4 °C in the dark. Cells were then washed with PBS/0.5% BSA, centrifuged at 800g for 2 min, followed by ice-cold PBS and incubated with Zombie NIR Fixable Viability dye (BioLegend, 423106) to stain dead cells for 20 min at 4 °C. After further washing the cells with PBS/0.5% BSA, cells were fixed and permeabilized in FOXP3 transcription factor fixation/permeabilization solution (Thermo Fisher Scientific) for 20 min at 4 °C, according to the manufacturer’s instructions. Intracellular antibodies were prepared in permeabilization buffer and incubated with cells for 30 min at 4 °C before cells were washed with permeabilization buffer, followed by PBS/0.5% BSA and finally resuspended in PBS/0.5% BSA. All of the experiments were performed using a five-laser BD LSRFortessa flow cytometer with DIVA software (BD Biosciences v.8.0.1). Compensation was determined automatically using Ultracomp eBeads (01-2222-42; Thermo Fisher Scientific). Data were analysed using FlowJo Software v.9.9.6.
Statistics and reproducibility
Statistical analyses were performed using GraphPad Prism, software (v9 GraphPad Software) and R (v.4.0.2 and higher) with statistical tests as indicated in the figure legends. Data were tested for normal distribution. All performed t-tests were two-tailed. P values are displayed in figures. No statistical methods were used to predetermine sample sizes but our sample sizes are similar to those reported in previous publications6,31,83,84,85. For animal experiments, biological replicate sizes were chosen taking into account the variability observed in pilot and previous studies in respective cohorts. For all experiments, animal/sample assignment was matched for age-matched control, and assigned based on randomly assigned mouse identification markings. Batched staining and analyses alongside controls were used throughout. The investigators were not blinded for the in vivo experiments. Technical staff administering therapy were blinded to the mouse genotypes. All subsequent tissue handling and analysis were blinded and/or performed using standardized automated analyses where possible. Quantitative image analysis was performed blinded to the genotype and treatment. The data distribution for normality testing and testing of equal variances was assessed using Prism 9 Software (GraphPad Software). No data were excluded, unless mentioned otherwise, except the following, which were excluded before analysis: one biological replicate failed quality control after transcriptomic sequencing—all other biological replicates from this and other cohorts successfully passed quality control and were included in downstream analysis; two drugs were excluded from the HCCO HTP screen due to microbiological contamination and drug precipitation in multiple replicates, respectively. One sample was excluded from the RFP expression analysis during analysis (total n = 4 biological replicates): testing AAV-mediated recombination of RFP alleles (Extended Data Fig. 1b,c), one sample was a notable outlier (4.9% versus 25.7%, 25.1% and 25.8%) which on re-review was caused by inconsistent RFP staining of the section—this outlier was removed from final analysis; details are provided in the figure legend also. Where the tumour number could not be quantified due to tumour rupture, no tumour number is reported (Fig. 5d).
Figures were assembled using Scribus v.1.4.8 (https://www.scribus.net/). Images were processed using Gimp v.2.10.14 (https://www.gimp.org/).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.