Human specimens
All human liver tissues used in this study were obtained after informed consent was obtained from patients undergoing operations at either the Department of Visceral, Thoracic and Vascular Surgery (VTG), University Hospital Carl Gustav Carus Dresden (UKD) or Leipzig University Medical Center. Informed consent was obtained from all participants. Use of the human samples for this study was approved by the corresponding institutional review boards of either the University Hospital Carl Gustav Carus Dresden (ethical vote BO-EK-57022020, ratified on 10 March 2020) or the Leipzig University Hospital (ethical vote: registration number 322/17-ek, date 10 June 2020 ratified 30 November 2021 and registration number 450/21-ek, date 21 November 2021 ratified on 4 October 2024). Five samples (F-PHH1–F-PHH5) were obtained from cryopreserved hepatocytes from Lonza Pharma&Biotech-Bioscience Solutions. Resected liver specimens were obtained from patients undergoing partial hepatectomy for benign or malignant conditions (for example, colorectal liver metastases, hepatocellular carcinoma or benign focal lesions). Only histologically normal, non-tumorous tissue adjacent to the resection site was used for organoid derivation. Clinical background information (sex, age, diagnosis/surgical indication) is provided in Supplementary Tables 1 and 2. Commercially obtained cryopreserved PHHs were derived from the livers of healthy donors deemed unsuitable for transplantation. Commercial number and supplier are given in Supplementary Table 2.
All procedures involving human material were conducted in accordance with the Declaration of Helsinki and institutional ethical guidelines.
Isolation of primary human hepatocytes and cholangiocytes
PHHs were isolated using a two-step collagenase perfusion method as described in refs. 34,61. The human liver tissue received from UKD was perfused with solution A (composed of 10 mM HEPES and 2.5 mM EGTA in HBSS) at 39 °C for at least 20 min, with a rate of 15 ml per 20 s. Subsequently, the perfusion solution was switched to solution B (containing 100 mM HEPES, 4.8 mM CaCl2 and 1 g l–1 collagenase P, in HBSS) and perfused at 37 °C for 5–15 min, also at a rate of 15 ml per 20 s. The digestion process was halted by adding cold William’s E medium supplemented with 1% HEPES, 1% GlutaMAX and 1% penicillin/streptomycin. PHHs were detached from the tissue by shaking using forceps and combing the cells out of the tissue. Afterwards, they were filtered through a 100-µm nylon cell strainer. Cells were then spun at 50g for 5 min, and the resulting pellet was resuspended in cold William’s E medium supplemented with 1% HEPES, 1% GlutaMAX and 1% penicillin/streptomycin. The cell suspension was kept cold and centrifuged again at 50g for 5 min.
For samples obtained from Leipzig University Hospital, the perfusion procedure differed slightly: solution A (composed of 10 mM HEPES (Carl Roth), 143 mM NaCl, 6.7 mM KCl, 2.4 mM EGTA, 5 mM N-acetyl-l-cysteine, 11 mM d-glucose (all provided by Sigma-Aldrich) and 32 U l–1 human insulin (Eli Lilly) in double-distilled water (pH 7.4)) at 39 °C with a rate of 25 ml per minute for at least 20 min. The perfusion solution was then switched to solution B (composed of 67 mM NaCl, 6.7 mM KCl, 10 mM HEPES, 0.5% BSA, 4.8 mM CaCl2 × 2H2O (all provided by Sigma-Aldrich), and 1 g l–1 collagenase P (Roche) in ddH2O (pH 7.6), diluted 1:2 in stop solution (composed of DPBS with Ca2+, Mg2+ (Gibco), supplemented with 16.7% FBS (Merck)) and perfused at 39 °C for 5–15 min at a rate of 25 ml min–1. The digestion was stopped by adding cold stop solution. Hepatocytes were filtered through a funnel with gauze (Hartmann) and centrifuged at 51g for 5 min. Cell pellets were washed in DPBS with Ca2+, Mg2+, centrifugated at 51g for 5 min and resuspended in William’s E medium supplemented with 10% FBS (Merck), 15 mM HEPES, 1 mM sodium pyruvate, 1% penicillin/streptomycin, 1% MEM NEAA (all provided by Gibco), 1 µg ml–1 dexamethasone (Jenapharm) and 32 U l–1 human insulin (Eli Lilly). The isolated PHHs were shipped overnight in ChillProtec plus medium (Biochrom).
Cryopreserved hepatocytes (F-PHH1–F-PHH5; Supplementary Table 2), commercially available from Lonza, were defrosted using human hepatocyte thawing medium (Lonza) following the manufacturer’s instructions.
The isolated PHH preparations (either from fresh tissue from Dresden or Leipzig Hospital or commercially available frozen hepatocytes) were enriched for both EpCAM-negative (hepatocytes) and EpCAM-positive (cholangiocytes) by MACS using an anti-human CD326 antibody (BioLegend) and anti-biotin microbeads (Ultra Pure, Miltenyi) following the manufacturer’s instructions. The EpCAM-negative fraction with a viability of >50% (Supplementary Table 1) was used to generate hepatocyte organoids as described below (see ‘Hepatocyte organoid culture’). The EpCAM-positive fraction, formed by human cholangiocytes, was used to generate h-CholOrgs as described previously4,5 and in ‘Cholangiocyte organoid culture’. A digestion method without perfusion, as the one detailed in ref. 4, only generated h-CholOrgs. h-HepOrgs were not formed under non-perfused protocols.
The complete list of patients used and the comparative between digestion and perfusion are provided in Supplementary Tables 1 and 2.
Flow cytometry validation of PHH purity following MACS enrichment
Freshly isolated PHHs and MACS-enriched EpCAM-negative PHHs (as described above) were centrifuged at 80g for 5 min. Pellets were resuspended in HBSS containing 2% FBS and incubated on ice for 10 min (blocking). After centrifugation (80g, 5 min), cells were resuspended in HBSS with 1% FBS, stained with EpCAM-Alexa 488 (5 μl per test; BioLegend), and incubated for 45 min on ice. Cells were then washed twice with HBSS containing 1% FBS, centrifuged and resuspended in 200 μl HBSS with 1% FBS, DAPI (1:1,000) and DNase I (1:1,000) for flow cytometry analysis.
Cholangiocyte organoid culture
For cholangiocyte organoid cultures, EpCAM-positive cholangiocytes were mixed with Matrigel growth factor reduced (Matrigel, Corning) or Cultrex basement membrane extract 2 (BME2) (Cultrex-RGF basement membrane extract type 2, BME2 (AMSBIO) at 50,000 cells per 50 μl in each well of a 24-well plate and cultured at 37 °C and 5% CO2 in h-CholOrg EM medium as described in refs. 4,5: AdDMEM/F12 medium containing 1% HEPES, 1% penicillin/streptomycin, 1% GlutaMAX, 1× B27 and 1.25 mM N-acetylcysteine (Sigma) supplemented with 10 nM gastrin (Merck/Sigma), 50 ng ml–1 hEGF (Peprotech), 10% RSPO1 conditioned medium (homemade), 100 ng ml–1 FGF10 (Peprotech), 10 mM nicotinamide (Merck/Sigma) and 25 ng ml–1 HGF (Peprotech)], 5 μM A8301 (Tocris) and 10 μM forskolin (Tocris, 1099). For the first 3–5 days in culture, this medium was supplemented with 30% WNT3a conditioned medium (Wnt-CM) (homemade), 25 ng ml–1 Noggin (Peprotech) and 10 μM ROCK inhibitor (Ri) (Y-27632, Merck/Sigma). The grown cholangiocyte organoids were passaged at a 1:3 ratio once a week as described in ref. 4. Organoid lines were routinely tested for mycoplasma.
Hepatocyte organoid culture
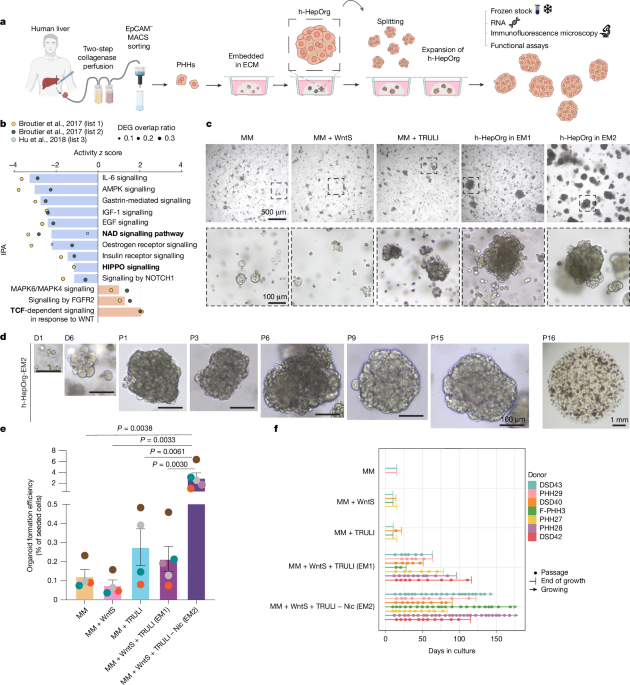
For hepatocyte organoid cultures, the isolated PHHs (EpCAM-negative fraction) were mixed with Matrigel (Corning) or BME2 (AMSBIO), and 12,500–50,000 cells were seeded in 50-μl domes per well in 24-well plates and incubated at 37 °C and 5% CO2. After gel solidification, culture medium was added. The culture medium was based on the medium from ref. 29 for hepatoblasts (MM) with modifications and the addition of WNT and YAP activation. The medium was composed of AdDMEM/F12 (Invitrogen) supplemented with 1% HEPES, 1% GlutaMAX (ThermoFisher), 1% penicillin/streptomycin (ThermoFisher), 1× B27 without retinoic acid (Gibco), 1.25 mM N-acetylcysteine (Sigma), 10 nM gastrin (Sigma) and the following growth factors: 50 ng ml–1 hEGF (Peprotech), 15% RSPO1 conditioned medium (home-made), 100 ng ml–1 FGF10 (Peprotech), 100 ng ml–1 FGF7 (Peprotech), 50 ng ml–1 HGF (Peprotech), 10 mM nicotinamide (Sigma, for EM1 medium only), 2 µM A83-01 (Tocris), 3 µM CHIR99021 (Tocris), 10 µM Y-27632 (Tocris), 0.5 nM Wnt surrogate Fc fusion protein as in ref. 40 (IPA, N001) and 10 µM TRULI (Axon) or 10 µM TDI-011536 (Selleckchem).
After 1 week to 10 days, the organoids were removed from the Matrigel or BME2, mechanically dissociated into small fragments using TrypLE Express (Gibco) and transferred to fresh Matrigel or BME2. Passaging was performed once per week at a 1:2 split ratio for at least 3 months. For preparation of frozen stocks, the organoid cultures were dissociated, mixed with Recovery cell culture freezing medium (Gibco) and frozen following standard procedures.
For the optimization of culture conditions, medium component screening experiments were performed in which each of the components Amphiregulin (AREG; 100 ng ml–1; R&D Systems), dexamethasone (1.6 µM; Sigma), G-CSF (50 ng ml–1; R&D Systems), IL-6 (2 ng ml–1; R&D Systems), M-3m3FBS (phospholipase C activator; 25 µM; Tocris), TGFα (100 ng ml–1) and TRULI (Axon) was added to our previously published mouse hepatoblast medium (MM29) with minor modifications: AdDMEM/F12 (Invitrogen) supplemented with 1% HEPES, 1% GlutaMAX, 1% penicillin/streptomycin, 1× B27 without retinoic acid, 1.25 mM N-acetylcysteine, 10 nM gastrin, 50 ng ml–1 hEGF, 15% RSPO1 conditioned medium, 100 ng ml–1 FGF10, 100 ng ml–1 FGF7, 50 ng ml–1 HGF, 10 mM nicotinamide, 2 µM A83-01, 3 µM CHIR99021, 10 µM Y-27632 and 0.5 nM Wnt surrogate Fc fusion protein. Note that addition of TRULI alone resulted in a significant increase in organoid formation efficiency (Fig. 1c,e). However, after 1–2 splits, the cultures rapidly deteriorated and could not be expanded further (Fig. 1f).
For h-HepOrg hepatic differentiation, h-HepOrgs were expanded in EM2 medium above, split, seeded and cultured for 2–5 days under EM1 culture medium, after which the medium was changed to DM medium composed of AdDMEM/F12 supplemented with 1% HEPES, 1% GlutaMAX, 1% penicillin/streptomycin, 1× B27 without retinoic acid, 1.25 mM N-acetylcysteine, 50 ng ml–1 hEGF, 15% RSPO1 conditioned medium, 50 ng ml–1 HGF, 2 µM A83-01, 3 µM CHIR99021, 10 µM Y-27632, 0.5 nM Wnt surrogate Fc fusion protein, 100 ng ml–1 FGF19 (R&D Systems) and 1.6 µM dexamethasone (Sigma). DM was changed every 2–3 days for 7 days.
For organoid formation efficiency, primary hepatocytes were isolated and cultured in different media as described above. To prevent organoids from fusing, 25,000 (for EM2 medium) or 50,000 (all other media) viable hepatocytes (viability of >80%) were plated in 50 μl Matrigel or BME2 and cultured as described above. After 12–14 days, organoid numbers were counted and results were expressed as a percentage relative to the initial seeding cell numbers. Organoid lines were routinely tested for mycoplasma.
Isolation of human liver portal fibroblasts
Human liver portal fibroblasts were isolated from human liver tissues by collagenase digestion. In brief, human liver tissue was minced and rinsed with cold DMEM (Gibco) supplemented with 1% HEPES, 1% GlutaMAX, 1% penicillin/streptomycin and 1% FBS. Minced tissues were incubated with a collagenase solution consisting of 2.5 mg ml–1 collagenase D (Roche), 0.1 mg ml–1 DNase I (Sigma), 1× B27 without retinoic acid, 1.25 mM N-acetylcysteine, 5% RSPO1 conditioned medium and 10 µM Y-27632 in DMEM supplemented with 1% HEPES, 1% GlutaMAX and 1% penicillin/streptomycin. Incubation was carried out for 30–60 min at 37 °C on a shaker set at 120 rpm. The digestion was halted by adding cold DMEM supplemented with 1% HEPES, 1% GlutaMAX, 1% penicillin/streptomycin and 1% FBS. The suspension was then filtered through a 70-µm cell strainer and centrifuged for 5 min at 300g. After removing the supernatant, the cell pellet was resuspended in cold DMEM supplemented with 1% HEPES, 1% GlutaMAX, 1% penicillin/streptomycin and 1% FBS. The suspension was centrifuged again for 5 min at 300g, and the resulting pellet was resuspended in cold DMEM supplemented with 1% HEPES, 1% GlutaMAX, 1% penicillin/streptomycin and 20% FBS. For sorting, portal fibroblasts were stained with anti-human CD90 (THY1) conjugated to APC, anti-human CD140a (PDGFRα) conjugated to PE, anti-CD11b/CD31/CD45 conjugated to PECy7 and anti-EpCAM conjugated to Alexa 488 for 30 min on ice and washed twice. THY1-positive portal fibroblasts were sorted using a BD FACSAria Fusion and cultured in DMEM supplemented with 1% HEPES, 1% GlutaMAX, 1% penicillin/streptomycin and 20% FBS at 37 °C and 5% CO2 until used for assembloid formation or frozen for biobanking. Portal fibroblast cultures were routinely tested for mycoplasma.
Viral infection
For portal fibroblast infections, cultures (passage 0 or 1) grown in DMEM+++ supplemented with 20% FBS (Merck/Sigma, F7524) were washed with PBS and dissociated to single cells by incubation with 1× TrypLE for 6 min at 37 °C. The cell concentration was determined by manual counting in a haemocytometer, and 10,000 cells were plated into each well of a 48-well plate and the medium mixed with nRFP- or nGFP-encoding lentivirus (LVP360-R and LVP360-G, GenTarget) to achieve a multiplicity of infection (MOI) of 10–30, then replaced after 12 h and the solution was changed after 72 h.
For cholangiocyte organoid infection, duct cells (passage 0 or 1) were extracted from Matrigel and digested with TrypLE to prepare single-cell suspensions as described in ref. 5, which were then manually counted using a haemocytometer to determine cell concentration. In a 48-well plate, 150 µl of cells and 50 µl of virus suspension from nRFP- or nGFP-encoding lentivirus (LVP360-R and LVP360-G, GenTarget) were added to achieve a MOI of 10–30, mixed thoroughly, centrifuged at 600g for 60 min at 32 °C and incubated for 6 h at 37 °C in 5% CO2. Cells were collected in 1.5-ml tubes and centrifuged at 600g for 5 min, the virus-containing medium was discarded and cells were resuspended in 25 µl of Matrigel, followed by the addition of cholangiocyte medium (supplemented with 30% WntCM, 25 ng ml–1 noggin and 10 µM Y-27632 for the first 3 days).
Periportal assembloids
To generate liver periportal assembloids comprising hepatocytes, cholangiocytes and portal fibroblasts, we first prepared the cellular components as follows: nGFP-labelled cholangiocyte organoids (passage 5–11), grown in cholangiocyte expansion medium (h-CholOrg-EM) as detailed above, were collected from Matrigel using cold AdDMEM/F12 (Invitrogen, 12634010) containing 1% HEPES (ThermoFisher, 15630-056), 1% penicillin/streptomycin (ThermoFisher, 15140-122) and 1% GlutaMAX (ThermoFisher, 35050038). Matrigel was removed and organoids were dissociated to single cells using prewarmed 1× TrypLE (Gibco) for 7–12 min at 37 °C. nRFP-labelled portal fibroblast cultures (passage 5–12) grown in DMEM+++ with 20% FBS (Merck/Sigma, F7524) were washed with PBS and dissociated to single cells by incubation with 1× TrypLE for 6 min at 37 °C. Both single-cell suspensions were spun at 200g for 5 min, resuspended in DM medium as described above but without A8301, and then manually counted with a haemocytometer to determine cell concentration. Cultured h-HepOrgs from EM2 medium were split and transferred to EM1 medium for 2 days and then to DM medium for 3 days. Hepatocyte organoids were then collected and washed using cold AdDMEM/F12 supplemented with 1% HEPES, 1% penicillin/streptomycin and 1% GlutaMAX and incubated for 10 min on ice using cold cell recovery solution (Corning, 354253) to remove the ECM. h-HepOrgs were then resuspended using DM without A8301 and placed into a low-attachment six-well plate; differentiated organoids (with bubbly morphology) were selected and hand-picked under a stereomicroscope.
To define an approach for human periportal liver assembloid formation, several iterations were performed. First, we sought to identify a medium that would support assembloid formation, that is, the culture of all three cell types: hepatocytes, cholangiocytes/ductal cells and portal mesenchyme without overgrowth of any of them, we tested several media and found that a minor adaptation of the DM medium used for h-HepOrgs differentiation without A8301 (assembloid medium) supported culture of the three cell types while preventing their overgrowth. To determine the optimal quantities of the three cell types required for periportal assembloid formation, we first investigated the proportions of portal fibroblasts and ductal cells in healthy human periportal liver tissue. We observed that the ratio varies from donor to donor from 1:1 to 4:1 ductal cells per fibroblast. Therefore, we tested this range of ratios in vitro by varying the proportions of mesenchyme and ductal cells that were mixed with a single h-HepOrg (~200-µm diameter). In short, in 96-well low-attachment U-bottom plates (Corning), we assembled (as described below) 1 h-HepOrg with 25 portal fibroblasts and 25, 50, 100 or 200 cholangiocytes, or with 100 cholangiocytes and 50 or 100 portal fibroblasts. We selected the proportion of 25 portal fibroblasts per 100 cholangiocytes/ductal cells. In AggreWell plates (AggreWell 800, Stem Cell Technologies), we scaled up proportionally, taking into account that the AggreWell 800 plate has 300 microwells in each well and used 7,500 portal fibroblasts, 30,000 cholangiocytes and 100 h-HepOrgs (proportion of 1 h-HepOrg to 75 portal fibroblasts and 300 cholangiocytes).
For non-healthy/non-physiological ratios, we used 500 portal fibroblasts, 100 cholangiocytes and 1 h-HepOrg for 96-well low-attachment U-bottom plates, and 15,0000 portal fibroblasts, 30,000 cholangiocytes and 50 h-HepOrgs for AggreWell plates.
For assembly in MW96, we mixed fibroblasts and cholangiocytes in 96-well low-adhesion U-bottom plates using 150 μl DM (without A8301) with 2.4 mg ml–1 methylcellulose (MeC; Sigma, M6385) and spun at 50g for 5 min. Individual h-HepOrgs were then added to the well and the mixture was incubated for 18–24 h at 37 °C and 5% CO2. For assembly in AggreWell plates, plates were first pretreated as recommended by the manufacturer. Then, ductal and mesenchymal cells together with h-HepOrgs were mixed in 1.5 ml DM (without A8301) with 2.4 mg ml–1 methylcellulose, spun down for 5 min at 50g and incubated for 18–24 h at 37 °C and 5% CO2. After 18–24 h in suspension in the 96-well/AggreWell plate, the cell suspension was collected with a 1-ml pipette and transferred to a low-attachment 6-well plate. The structures were manually picked under a stereomicroscope and seeded in 25 μl Matrigel dome in prewarmed 48-well plates. The Matrigel was allowed to solidify for 30 min at 37 °C in 5% CO2, and the wells were overlayed with an additional 300 μl of DM (without A8301). The medium was changed every 3–4 days. Under these conditions, 70% of the initial cholangiocytes formed a lumen. Raw data were incorporated into the quantification of periportal-like spatial organization in assembloids (source data for Extended Data Fig. 7e).
Immunostaining of organoids and assembloids
For immunofluorescence staining, organoids and assembloids were first extracted from Matrigel with ice-cold Cell Recovery solution and then fixed for 30 min with 4% paraformaldehyde (PFA) at 4 °C. Fixed organoids were washed and transferred to µ-Slide 8-well chamber slides (glass bottom; Ibidi). Blocking and permeabilization were performed for 1 h at room temperature in PBS containing 2% BSA and 0.1%, 0.2%, 0.5% or 1% Triton X-100 depending on the antigen (Supplementary Data 5). The samples were incubated with primary antibodies overnight at 4 °C in blocking solution. After that, the antibody was washed with three washes with PBS and the samples were incubated overnight at 4 °C or for 8 h at room temperature with secondary antibodies diluted in blocking solution and, if required, also phalloidin and DAPI were added to the secondary antibody mix. The samples were washed three times with PBS and subsequently cleared using fructose-glycerol clearing solution (25 ml glycerol, 5.3 ml dH2O and 22.5 g fructose–60% glycerol and 2.5 M fructose). The samples were stored in PBS until they were cleared for imaging as described above. The antibodies and dilutions used are listed in Supplementary Data 5.
For haematoxylin and eosin (H&E) staining, organoids were collected in cold DPBS (Gibco) and fixed with 4% PFA for 30 min and dehydrated and embedded in paraffin using standard methods. Paraffin sections (8 μm) were cut and stained for H&E using standard protocols.
Immunostaining of thin and thick tissue sections
For thin tissue sections (8–12 μm) and staining, human liver tissues were fixed in 10% formalin overnight with rolling at 4 °C. After fixation, tissues were washed with PBS and incubated with 10% sucrose for 1–2 h, then transferred to 30% sucrose in PBS for 24 h and subsequently embedded in OCT compound (VWR, 361603E) to generate OCT cryopreserved tissue blocks. Tissue blocks were cryosectioned on a CryoStar NX70 cryostat (ThermoScientific). Sections were blocked in PBS with 10% donkey serum (DS) and 0.1% Triton X-100 for 2 h at room temperature, incubated with primary antibodies diluted in PBS with 3% donkey serum and 0.1% Triton X-100 overnight at 4 °C and subsequently washed and incubated with secondary antibodies diluted in 0.05% BSA in PBS and DAPI for 2 h at room temperature. Sections were mounted in Vectashield. The list of antibodies used is available in Supplementary Data 5.
For thick tissue sections and staining, the protocol from ref. 62 was used. Immediately after surgical resection, liver tissue samples were cut into smaller pieces and fixed in 4% PFA for 24 h on a rotator at 4 °C and washed three times with PBS, followed by quenching with 50 mM ammonium chloride solution (NH4Cl) for 24 h and again washed three times with PBS. For storage, liver pieces were kept in PBS at 4 °C. For sectioning, livers were embedded in moulds with 4% low-melting agarose (Bio-Rad, 1613111) in PBS and cut into 50- or 100-μm-thick sections on a vibratome (Leica, VT1200S). For deep tissue imaging, if antigen retrieval was required, tissue sections were placed in Eppendorf tubes with prewarmed 1× citrate buffer (Sigma-Aldrich, C9999), pH 6, at 80 °C for 30 min in a shaking heating block and then washed three times with PBS. Tissue sections were permeabilized with 0.5% Triton X-100 in PBS for 1 h at room temperature. The primary antibodies were diluted in Tx buffer (0.2% gelatin, 300 mM NaCl and 0.3% Triton X-100 in PBS) and incubated for 48 h at room temperature. After washing three times for 15 min each with 0.3% Triton X-100 in PBS, the sections were incubated with secondary antibodies, DAPI (1 mg ml–1; 1:1,000) and phalloidin for another 48 h. After washing three times for 15 min each with 0.3% Triton X-100 in PBS and three times for 1 min each with PBS, the optical clearing started by incubating the slices in 25% fructose for 4 h, continued in 50% fructose for 4 h, 70% fructose overnight, 100% fructose (100% wt/vol fructose, 0.5% 1-thioglycerol and 0.1 M phosphate buffer, pH 7.5) overnight, followed by a final overnight incubation in SeeDB solution (80.2% (wt/wt) fructose, 0.5% 1-thioglycerol and 0.1 M phosphate buffer)63. The samples were mounted in SeeDB. A list of antibodies and dyes used is available in Supplementary Data 5.
For immunohistochemistry of tissue sections from xenotransplanted mice, mouse liver tissue samples were cut into smaller pieces and fixed in 10% formalin overnight. Sections (4 μm) were subjected to immunohistochemical staining, which was performed using a Dako REAL EnVision detection system (Dako, K5007). Anti-human GAPDH antibody (Abcam) (Supplementary Data 5) was used as the primary antibody and nuclei were counterstained with haematoxylin. Stained tissues were viewed under a Virtual Slide System (Leica, ScanScope CS2).
The immunohistochemistry analysis for PDGFRA, DCN and ASPN in healthy human liver tissue was obtained from the publicly available image dataset from Human Protein Atlas (HPA)64 (version 24proteinatlas.org). The corresponding URL is indicated in the figure legend.
Imaging of organoids, assembloids and tissues
Bright-field images of organoids were obtained with a Leica DMIL LED inverted microscope and Leica DFC 450C camera or with a Leica M80 stereoscope and MC170HD camera and Leica LAS software. H&E staining of organoids was obtained with a Leica DM4B microscope and DMC5400 camera and Leica LAS X software.
Confocal images of organoids and thick tissue sections were acquired on an inverted single-photon point scanning confocal microscope (Zeiss Cell Discoverer 7 with LSM 900 and Airyscan 2) using a Zeiss APOCHROMAT ×20/0.95-NA Autocorr air objective, with a tube lens of ×0.5 or ×1, and a voxel size of 0.4 × 0.4 × 0.5 μm or 0.5 × 0.5 × 0.5 μm for organoids and 0.3 × 0.3 × 0.3 μm for thick tissue sections. Laser lines 405, 488, 561 and 640 were used for excitation of fluorophores, and GaAsP-PMT detectors were used for detection. High-resolution Airyscan images were acquired using this system for imaging polarity in detail for the tissue sections with a voxel size of 0.0823 × 0.0823 × 0.3 μm. Image processing was done using Zen software or ImageJ/Fiji.
Imaging of assembloids and thin tissue sections was performed using an inverted multiphoton laser-scanning microscope (Zeiss LSM 780 NLO). To improve the resolution, image denoising was performed with deconvolution using HuygensPro. Raw image stacks were imported into the software, and a point spread function (PSF) was either estimated based on the imaging conditions (numerical aperture, wavelength and refractive index) or obtained from PSF calibration images. The HuygensPro classic maximum likelihood estimation (CMLE) algorithm was applied for deconvolution, with an iteration stop criterion based on optimal signal-to-noise ratio and minimal change in successive iterations.
Image analysis
Quantification of the percentage of YAP-positive and YAP-negative nuclei was performed using Arivis 4D Pro software (version 4.2.0). The steps of the analysis pipeline included background correction, denoising, nuclear segmentation based on DAPI and quantification of the fluorescence intensity of YAP immunofluorescent staining in the nuclei. The total number of nuclei and the number of YAP-positive nuclei were quantified, and, subsequently, the number of YAP-negative nuclei was calculated by subtracting the number of YAP-positive nuclei from the total number of nuclei. Finally, the percentages of YAP-positive and YAP-negative nuclei were calculated.
Quantification of cytoplasmic to nuclear area was performed using Arivis 4D Pro software (version 4.2.0). For this, a representative 2D z slice was taken from each organoid. The analysis pipeline included preprocessing steps of background correction on the phalloidin channel (marking cell borders) and normalization and denoising on the DAPI channel (marking nuclei). To obtain the nuclear area, nuclear segmentation was done based on DAPI, followed by quantification of the total nuclear area. For the cytoplasmic area, segmentation was done based on phalloidin to obtain the outline of the area occupied by the cytoplasm. Finally, the ratio of cytoplasmic area to nuclear area was calculated.
For 3D visualization of bile canaliculi, high-resolution images were obtained as described above. Segmentation was performed on CD13 (for bile canaliculi) and F-actin (cell borders) staining with phalloidin. Analysis of bile canaliculus morphology and bile canaliculus network properties was performed using a custom-made Fiji script publicly available at https://git.mpi-cbg.de/huch_lab/assembloid-paper. A description of the script can be found in ref. 51 In brief, immunofluorescence images from several conditions were used in this analysis: EM2, DM and liver tissue, from hereon referred as ‘structure’. We refer to individual bile canaliculus networks as ‘network’. We determined the connectivity of the network by analysing the total number of branching points (number of triple junctions) per structure. We determined the length of the network per structure by analysing the total length of all branches in the structure. To compare structures in different conditions, we plotted these values as dot plots in which each dot was one structure. In the case of tissue, each dot was one field of view. The features extracted from Fiji were exported as .csv files and plotted using Prism.
For assembloids, to visualize the structure from different angles, immunofluorescence images were visualized in 3D using MotionTracking (http://motiontracking.mpi-cbg.de)43. For this, Gaussian blurring was applied to the channels of interest and then visualized in 3D.
For quantification of cholangiocytes and portal fibroblasts in assembloids, Arivis 4D software (Zeiss) was used. For the analysis, nuclei were segmented based on diameter, probability threshold and split sensitivity to align with the expected morphology in the fluorescence images. When segmentation was incomplete due to weak fluorescence signals, missing nuclei were manually added. This approach was used to determine the number of nuclei per cell and the number of cells per organoid. All segmentation results were manually reviewed and corrected as necessary.
Isolation of mRNA and RT–qPCR analysis
RNA was extracted from organoid cultures or freshly isolated tissue using the RNeasy Mini RNA Extraction Kit (Qiagen) with DNase treatment and reverse-transcribed using Moloney murine leukaemia virus reverse transcriptase (Promega). All targets were amplified (40 cycles) using gene-specific primers (Key Resource Table) and PowerUp SYBR Green master mix (ThermoFisher) or iQ SYBR Green Supermix (Bio-Rad) and run on a qPCR instrument (Thermo Fisher QuantStudio 7 Pro or GeneAmp PCR System 9700; Applied Biosystems respectively). Data were analysed using Design & Analysis 2.7.0 software (ThermoFisher).
Karyotyping
Mitotic metaphases for karyotyping were obtained by subculturing hepatocyte organoids in the active growth phase. The following day, cells were exposed to 0.2 μg ml–1 colcemid (Gibco) for 60 min at 37 °C to arrest them in metaphase. Organoids were dissociated into single cells using TrypLE Express (Gibco). After centrifugation and removal of the supernatant, cells were subjected to hypotonic treatment with a solution of 0.075 M KCl for 30 min at 37 °C, followed by fixation in a 3:1 methanol to acetic acid solution. The preparation was washed three times with the fixative before slide preparation. Chromosomes were stained with Giemsa (Merck) diluted in Gurr buffer (pH 6.8; Gibco). Images were taken with a Zeiss Axio Imager.Z2 upright motorized stand with an ApoTome.2 for improved z contrast.
Functional assays
For functional assays, h-HepOrgs were cultured in EM and DM media and assembloids in DM media as described above. As negative controls, we used h-CholOrgs grown as described above. As positive controls, we used freshly isolated PHHs cultured in standard 2D hepatocyte monolayer culture or in sandwich culture65. In brief, for the positive control of 2D hepatocytes, fresh isolated PHHs were plated onto collagen (1.8 mg ml–1; RatCol collagen, Advanced Biomatrix)-coated 24-well plates at 500,000 or 250,000 cells per well in William’s E medium (PAN Biotech) supplemented with 10% FBS, penicillin/streptomycin and 100 nM dexamethasone for 3 h for attachment. For the monolayer culture (1d-PHH monolayer control), the cells were cultured on William’s E medium supplemented with 1% HEPES + 1% GlutaMAX + 1% penicillin/streptomycin and 100 nM dexamethasone for 18 h (or 24 h, for albumin assays) and then processed for the functional assays. For sandwich cultures, fresh isolated PHHs were plated onto collagen as above and overlayed with a second collagen layer (1.2 mg ml–1; RatCol collagen, Advanced Biomatrix) and cultured for 7 days in William’s E medium supplemented with CM4000 cell maintenance supplement (ThermoFisher Scientific).
To determine albumin secretion, supernatant from 24 h was collected and the amount of albumin was determined using a human-specific albumin ELISA kit (Assay Pro) following the manufacturer’s instructions on an ELISA plate reader (Tecan Spark 20M). To measure cytochrome P450 activity, on the day of the experiment cholangiocyte and hepatocyte organoids in EM2 or DM were removed from Matrigel using Cell Recovery solution (Corning). Organoids, 2D hepatocyte monolayers or 2D sandwich cultures were then all cultured in William’s E medium supplemented with 1% HEPES + 1% GlutaMAX + 1% penicillin/streptomycin supplemented with luciferin-H substrate (100 µM) or luciferin-IPA (3 µM) for 6 h. Cytochrome activity was measured using the P450-Glo Assay Kit (Promega) according to the manufacturer’s instructions on a plate reader (PerkinElmer Envision). Results were normalized to total viable cell counts per well.
Urea synthesis assay
To determine urea secretion, cell culture supernatants were collected from 48-well plate after 12 h of culture. The concentration of secreted urea was measured by Urea Assay Kit (Abnova) according to the manufacturer’s instructions.
Measurement of gluconeogenesis
Gluconeogenesis was assessed using a Glucose-Glo Assay (Promega). Organoids/assembloids were first washed twice with PBS to remove residual glucose and then incubated for 24 h in glucose-free medium (Gibco) to deplete intracellular glucose stores. Subsequently, the organoids were stimulated for 24 h in gluconeogenesis-inducing medium (glucose-free medium supplemented with 10 mM lactate; Sigma-Aldrich, L7022) to promote hepatic glucose production.
After incubation, 25 µl of supernatant from each well was transferred to a 96-well assay plate and mixed with an equal volume of glucose detection reagent. Following incubation for 60 min at 37 °C, luminescence was measured using a luminometer.
Cell counting
h-HepOrgs were dissociated into single cells using 10× TrypLE (Gibco) after 10 and 15 days of culture in specified media. Cell counts were determined using a Countess II FL automated cell counter (ThermoFisher Scientific).
Quantification of xenobiotic metabolism by mass spectrometry
h-HepOrgs were cultured in DM as previously described. Assembloids were maintained under the same conditions for 6 days. Freshly isolated PHHs were cultured in a monolayer for 24 h, also as described above. Following culture, all cells were washed twice with PBS. The medium was then replaced with 100 μl of William’s E medium supplemented with 1% HEPES, 1% GlutaMAX, 1% penicillin/streptomycin and verapamil (Merck) at a final concentration of 4 µM. Cells were incubated for 6 h, after which the supernatant was collected and analysed by mass spectrometry.
Organoids and assembloids were dissociated into single cells using 10× TrypLE and manually counted using a haemocytometer. The resulting cells were washed twice with PBS and stored at –20 °C.
Metabolites were separately extracted from the supernatant and from the cells by isopropanol:methanol:chloroform mixture (4:2:1, v/v/v) containing 7.5 mM ammonium formate (termed MS mix). A supernatant aliquot of 100 μl was diluted 20-fold (v/v) with MS mix, vortexed, centrifuged for 7 min at 13g and the pellet was discarded. Cells suspended in 100 μl PBS were first lysed using ~25 stainless steel beads of 0.5 mm in size (Next Advance, USA, 152034) in the Qiagen Ratsch Tissue Lyser at 30 Hz for 8 min and metabolites were extracted as above. Each sample was prepared in three biological replicates and analysed by mass spectrometry immediately after extraction.
Mass spectrometry analysis was performed on a Q Exactive hybrid quadrupole Orbitrap tandem mass spectrometer (ThermoFisher Scientific) in positive ion mode by direct infusion of total extracts. Prior analyses, the internal standard verapamil-13C3 hydrochloride (Merck) was dissolved in methanol and spiked into samples to a final concentration of 200 nM. Aliquots of 40 μl of each sample were then placed on twin.tech PCR Plate 96 (Eppendorf, 0030128.648) and infused into the mass spectrometer via TriVersa NanoMate robotic ion source (Advion Interchim Scientific) using nanoflow chips with a nozzle diameter of 4.1 μm. The ion source was controlled using Chipsoft 8.1.0 software. Spraying voltage and gas back pressure were set to 1.25 kV and 0.95 psi, respectively. The ion transfer capillary temperature was set to 200 °C and the S-lens RF level was set to 50%. A target mass resolution (Rm/z) of 200 was set to 140,000 (full width at half maximum, FWHM) for both Fourier transform mass spectrometry (FT MS) and FT MS/MS spectra. To acquire FT MS spectra, the automated gain control (AGC) was set to 3 × 106, the maximum injection time was set to 500 ms, the acquired mass range m/z was 50–700, the lock masses m/z 445.12003 and m/z 338.34174. The acquisition cycle consisted of recording FT MS1 spectra for 1.2 min followed by two FT MS/MS2 spectra for 1.8 min from the precursors with m/z 455.291 (for verapamil [M + H]+) and m/z 441.275 (for norverapamil [M + H]+); precursor m/z isolation width was 3 Th.
Spectra were averaged in Xcalibur Qual Browser v.3.0 (ThermoFisher Scientific) over a 30-s time range corresponding to stable spray; peaks of metabolites and standard extracted with 5 ppm mass accuracy. The absolute amount of norverapamil was calculated from its molecular ion intensity normalized to the intensity of the standard. For calibration, aliquots of William’s E medium containing verapamil (Merck, V-002-1ML) with the concentration ranging from 2 μM to 8 nM were diluted 20-fold with MS mix, spiked with the internal standard and analysed as described above. The determined abundance of norverapamil in supernatant and in cellular pellets was summed up, normalized to 104 cells and its production rate was expressed in pmol/h.
Xenotransplantation in Fah
−/−
Rag2
−/−
Il2rg
−/− (FRG) mice
Male and female Fah−/−Rag2−/−Il2rg−/− (FRG) mice were obtained from Jackson Laboratory. Mice were housed and maintained under specific-pathogen-free conditions in accordance with the principles of laboratory animal care and the guide set by the HYU Industry-University Cooperation Foundation. All animal experiments were conducted under protocols approved by the Institutional Animal Care and Use Committee (IACUC) of Hanyang University (2024-0148B). Experimental groups were not predetermined based on the sex of the mice, and all animals were randomly assigned to experimental procedures. Male mice accounted for approximately 25% of the total cohort. FRG mice 8–16 weeks old were used for all experiments. For their maintenance, mice were administered ad libitum NTBC (2-(2-nitro-4-trifluoromethylbenzoyl)-1,3-cyclohexanedione) in their drinking water.
Mice aged 8–16 weeks of both sexes were kept on NTBC in drinking water until 3 days before the experiment, when NTBC was withdrawn. h-HepOrgs expanded in EM2 and differentiated in DM were dissociated into single cells and prepared for injection. For transplantation experiments, commercially available frozen PHHs were used (F-PHH2; Supplementary Table 2). Organoids cultured under EM2 medium as well as isolated hepatocytes (PHHs) from the same donors were used as controls. Following dissociation, 500,000 dissociated organoid cells or 800,000 PHHs were resuspended in 100 μl AdDMEM/F-12 medium and injected into the spleen. The non-injected negative-control group received 100 μl PBS instead of cells. Mice were cycled in and out of NTBC treatment for 3 days every time their body weight dropped below 80% of the initial weight.
IPA
We performed IPA (Qiagen) to identify potential candidate signalling pathways. For this, we first generated three DEG lists as DEGs between liver cancer organoids and liver healthy (list 1) or cancer (list 2) tissue (Supplementary Data 1_S1) and DEG list between partial hepatectomy and healthy tissue (list 3). Gene lists were generated as follows: lists 1 and 2: gene expression matrices from hepatocellular carcinoma (HCC)-derived organoids, HCC liver tissue and liver tissue from healthy donors were obtained from the Gene Expression Omnibus (GEO) under accession number GSE84073 (ref. 35). DEGs were identified using DESeq2 (ref. 2), applying a threshold of |log2 fold change| > 1 and an adjusted P value of <0.1 (Supplementary Data 1_S1). For list 3, DEGs comparing partial hepatectomy and undamaged liver hepatocytes in mouse were sourced from the supplementary tables in ref. 27. Additionally, a list of genes mutated in both HCC-derived organoid lines was derived from the whole-exome sequencing (WES) results in ref. 35 (list 4). The full list of DEGs from lists 1–4 is provided in Supplementary Data 1_S1.
The three DEG lists and the mutated gene list (lists 1–4) were analysed using IPA, using the canonical pathway analysis and upstream regulator prediction functions (QIAGEN Inc., https://digitalinsights.qiagen.com/ipa). In brief, the significance of the association between the dataset and canonical pathways was determined using a right-tailed Fisher’s exact test, followed by Benjamini–Hochberg correction for multiple testing. For analyses in which log fold changes were available, an activity z score was computed to predict the activation or inhibition likelihood of specific pathways base. Upstream regulator analysis used a computational algorithm to identify upstream regulators potentially responsible for the observed gene expression changes. From the IPA canonical pathway analysis, pathways were filtered based on an adjusted P value of <0.05 and the presence of the keyword ‘signalling’ in the pathway name (Supplementary Data 1_S2). Selected pathways of interest with a mean adjusted P value and frequency of pathway significance across comparisons are plotted in Extended Data Fig. 1c (Supplementary Data 1_S3). Activity z scores from the selected pathways were individually plotted as well as their corresponding mean values in Fig. 1b (Supplementary Data 1_S4,5). Next, results from the upstream regulator analysis were filtered for (1) an adjusted P value of <0.1 as upstream regulator and (2) the molecules from the two selected signalling pathways (Supplementary Data 1_S6). Key components of the signalling pathways and their adjusted P value in upstream regulator analysis are plotted in Extended Data Fig. 1d (Supplementary data 1_S7).
Bulk RNA-seq library preparation
mRNA was isolated from on average 270 ng total RNA by poly(dT) enrichment using the NEBNext Poly(A) mRNA Magnetic Isolation Module (NEB) according to the manufacturer’s instructions. Samples were then directly subjected to the workflow for strand-specific RNA-seq library preparation (Ultra II Directional RNA Library Prep, NEB). For ligation, NEB Next Adapter for Illumina of the NEB Next Multiplex Oligos for Illumina Kit was used. After ligation, adapters were depleted by XP bead purification (BeckmanCoulter) adding the bead solution in a ratio of 0.9:1 to the samples. Unique dual indexing was done during the following PCR enrichment (12 cycles) using amplification primers carrying the same sequence for i7 and i5 index (i5: AATGATACGGCGACCACCGAGATCTACACNNNNNNNNACATCTTTCCCTACACGACGCTCTTCCGATCT; i7: CAAGCAGAAGACGGCATACGAGATNNNNNNNNGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT). After two more XP bead purification steps (0.9:1), libraries were quantified using the Fragment Analyzer (Agilent). Libraries were sequenced on an Illumina NovaSeq 6000 in 100-bp paired-end mode to a depth of 40 million read pairs per library.
RNA-seq data processing
Raw bulk RNA-seq data were processed using nf-core/rnaseq v3.18.0 (https://doi.org/10.5281/zenodo.1400710) of the nf-core collection of workflows66, using reproducible software environments from the Bioconda67 and Biocontainers68 projects. The pipeline was executed with Nextflow (v24.10.5)69. The reference genome used was Homo sapiens GRCh38 (Ensembl release 111). The pipeline was run with custom parameters for trimming (extra_trimgalore_args: ‘–nextseq 20 –length 15’), alignment (extra_star_align_args: ‘–outFilterMismatchNmax 999 –outFilterMismatchNoverLmax 0.1 –alignMatesGapMax 200000 –chimSegmentMin 20 –twopassMode Basic –alignIntronMin 20 –alignIntronMax 200000’) and quantification (extra_salmon_quant_args: ‘–seqBias –gcBias –posBias’). The resulting MultiQC report was inspected to ensure overall sequencing quality and pipeline performance.
Transcript-level abundance estimates were imported using the tximeta package70 to generate a gene-level count matrix. Next, variance stabilizing transformation (VST) from DESeq2 (refs. 71,72) was used to normalize the data. Euclidean distance matrices, principal-component analysis (PCA) and heat map visualizations were computed on the VST-transformed values. On some heat maps, minimum–maximum scaling was applied. In Extended Data Fig. 2a,b, batch correction was performed on the VST-transformed values using limma’s removeBatchEffect, with sample material type (tissue versus organoid) treated as the batch variable73. For differential expression analysis, DESeq2 was used. For comparison between MM + WntS + TRULI and primary (fresh isolated PHHs), the design formula ~ donor + condition_l3 was applied (Extended Data Fig. 2). Log-fold changes were shrunken using lfcShrink with the ashr method (type = ‘ashr’), applying a fold-change threshold of 1.5 and a significance threshold of α = 0.05 (ref. 74). For the comparison between DM and EM2 (Fig. 2e), the design formula ~ batch + donor + condition_l1 was applied. Log-fold changes were shrunken using lfcShrink with the ashr method (type = ‘ashr’), applying a fold-change threshold of 1.5 and a significance threshold of α = 0.05. For the comparison between h-HepOrgs and portal fibroblasts (Extended Data Fig. 6h), the design formula ~sex + cell_type was applied. Log-fold changes were shrunken using lfcShrink with the ashr method (type = ‘ashr’), applying a fold-change threshold of 4 and a significance threshold of α = 0.05. Gene set enrichment analysis (GSEA) was conducted using the clusterProfiler package, leveraging gseKEGG, gseGO and gsePathway for pathway enrichment analysis75.
The zonated gene list (Extended Data Fig. 3h) was obtained by manually curating genes that have been confirmed to be portally or centrally zonated from human spatial transcriptomic datasets10,11,46,47 (a full list is provided in Supplementary Data 2_S6). We then intersected this refined zonated gene list with our list of differentially expressed genes in the DM versus EM2 comparison.
Donor-specific genes were identified separately for batches Y1/Y2 and S1/S2 using a likelihood ratio test (LRT) with the full model ~donor and the reduced model ~1. Genes with an adjusted P value of <0.05 were retained, and the resulting gene lists from the two batches were merged. Pairwise correlations between organoids and primary cells were computed using the donor-specific genes. For the heat map shown in Extended Data Fig. 4e, sex-specific genes were excluded.
The complete software stack for downstream analysis is available as a Docker container (rnaseq-notebook:2025-04-21) archived at https://quay.io/repository/fbnrst/rnaseq-notebook and archived on Zenodo (https://doi.org/10.5281/zenodo.17704466).
Single-cell transcriptomics with 10x Genomics
For scRNA-seq analysis, assembloids were generated by assembling h-HepOrgs, cholangiocytes/ductal cells derived from cholangiocyte organoids (n-GFP) and portal fibroblasts (n-RFP) at a ratio of 1 h-HepOrg to 25 portal fibroblasts and 100 cholangiocytes. At 5–6 days after aggregation, assembloids were collected as follows: periportal assembloids were dissociated to single cells using 10× TrypLE for 5 min at 37 °C. The cells were resuspended in DM and 10 μg ml–1 DNase in BSA-coated tubes and filtered through a 100-μm strainer. Cell suspensions (30,000–50,000 cells) were concentrated by centrifugation (50g, 5 min, 4 °C) and the volume was reduced to ~55 µl. Cells were carefully resuspended and visually inspected under a light microscope to determine cell concentration and quality. The concentrations of the single-cell suspensions were adjusted to 138–912 cells per microliter and carefully mixed with the reverse transcription mix before loading cells on the 10x Genomics Chromium system76 in a Chromium Single-Cell G Chip targeting 3,000–10,000 cells per reaction. Following the guidelines of the 10x Genomics Chromium Single-Cell Kit v3.1 user manual, the droplets were directly subjected to reverse transcription, the emulsion was broken and cDNA was purified using Dynabeads MyOne Silane (10x Genomics). cDNA was first amplified with 12 cycles, and then purified with 0.6× SPRIselect beads (BeckmanCoulter) to enrich cDNA fragments (>400 bp). A quality and quantity control of cDNA on the Fragment Analyzer (using the DNF-473 NGS Fragment Kit, Agilent) was eventually performed to obtain its concentration. The 10x Genomics scRNA-seq library preparation–involving fragmentation, dA tailing, adapter ligation and 11 or 12 cycles of indexing PCR, was performed based on the manufacturer’s protocol. After quantification, the libraries were sequenced on an Illumina NovaSeq 6000 in paired-end mode (R1/R2, 100 cycles; I1/I2, 10 cycles), generating 230–370 million fragment pairs.
scRNA-seq data analysis
The raw scRNA-seq data were processed using nf-core/scrnaseq v3.0.0 (https://doi.org/10.5281/zenodo.3568187) of the nf-core collection of workflows66, using reproducible software environments from the Bioconda67 and Biocontainers68 projects. The pipeline was executed with Nextflow (v24.10.5)69. STARSOLO was used as the aligner. The reference genome was set to Homo sapiens GRCh38 (Ensembl release 111) with custom additions for RFP and GFP transgenes, obtained from SnapGene (DsRed1 and EGFP, respectively). Outputs were inspected for quality control, and one sample with poor quality control was excluded from further analysis. Within nf-core/scrnaseq, technical artefacts were eliminated using CellBender77. The CellBender output was used for data visualization. Doublet detection for each sample was performed using scrublet78.
Further analysis was performed using scanpy79. Quality control was applied with the following thresholds: minimum total counts of 5,000, minimum detected genes of 2,000, the maximum percentage of counts in the top 50 genes set at 50%, the maximum percentage of mitochondrial counts set at 15% and a maximum doublet score of 0.15. Gene filtering was performed to retain genes expressed in at least ten cells. After filtering, the data underwent normalization, log transformation and identification of the top 3,000 most highly variable genes. PCA was performed, and batch correction was implemented through Harmony integration80. UMAP visualization and Leiden clustering were used to identify the three expected cell types81,82.
To compare homeostatic-like and fibrotic-like organoids, pseudobulk aggregation was performed using decoupleR for each cell type83. Pseudobulk data were generated by summing raw counts for each sample and cell type, with a minimum requirement of ten cells per group and 1,000 total counts. Differential expression analysis was conducted using pyDESeq2 (ref. 84). For each cell type, DESeq2 datasets were created with design factors that included ‘donor’ and ‘condition’, using the ‘homeostatic-like’ condition as the reference. Differentially expressed genes between the homeostatic-like and fibrotic-like conditions were ranked on the basis of the test statistic. Subsequently, gene set enrichment analysis (GSEA) was performed on the ranked lists using clusterProfiler, focusing on KEGG, Reactome and GO terms.
The complete software stack for downstream analysis is available as a Docker container (singlecell-notebook:2025-04-21) archived at https://quay.io/repository/fbnrst/singlecell-notebook and archived on Zenodo (https://doi.org/10.5281/zenodo.17704461).
Comparison to public datasets
Data from refs. 12,13,49 were downloaded in h5ad format from https://cellxgene.cziscience.com/. Additionally, data from ref. 11 were obtained from https://data.mendeley.com/datasets/yp3txzw64c/1, and the dataset from ref. 8 was downloaded from https://datashare.ed.ac.uk/bitstream/handle/10283/3433/tissue.rdata and converted to h5ad format using the sceasy package.
These public datasets were merged with the raw count matrix of our quality control-filtered organoid data. Subsequently, the combined dataset underwent normalization, followed by log transformation and detection of the top 4,000 most highly variable genes. We performed PCA and integrated the dataset using Harmony, specifying concatenation of the paper and donor as batch variables, with a maximum of 20 iterations and a theta value of 1.5. Selected genes were visualized in a dot plot (Fig. 4h).
Pseudobulk analyses were then conducted using the decoupleR package to summarize gene expression by cell type. This involved generating a pseudobulk dataset in which raw counts were summed by sample and cell type, ensuring a minimum of 30 cells per group. Following pseudobulk aggregation, the data were normalized and log transformed, with the top most highly variable genes identified on the basis of mean expression and dispersion. Additionally, the ‘paper’ variable was regressed out to mitigate batch effects. Next, PCA was performed on the pseudobulk data, using 50 principal components for subsequent analyses. Hierarchical clustering was executed using the Pearson correlation metric, and Pearson correlation matrices were plotted, as shown in Figs. 4g and 5f.
Marker genes for the three major cell types were computed separately for our organoid data and the merged public data using scanpy’s rank_genes_groups function. For each dataset, the top 300 marker genes for each cell type were selected. Subsequently, GSEA was performed using the gseapy package, leveraging the Enrichr method85. The analysis focused on the KEGG 2021 Human and Reactome 2022 gene sets, with a P-value cut-off of 0.05. Shared enriched pathways between the organoid and tissue datasets were identified, and the combined enrichment scores for selected terms were plotted (Extended Data Fig. 8c,d).
Data statistical analysis
The specific statistical test is specified in the legend. P < 0.05 was considered statistically significant. In all cases, data from at least three independent experiments were used. Calculations were performed using the Prism 9 software package. All P values are given in the corresponding figure legends or in the corresponding figure or in the corresponding source data file. Dispersion and precision measures (for example, mean, median, s.d., s.e.m.) are specified in the figure legends. No statistical methods were used to predetermine sample size. All scRNA-seq statistics are described above in the corresponding section.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.