Human specimens
Human liver tissues for the single-nucleus sequencing were obtained from different sources as shown in Supplementary Table 10. Liver samples were collected under IRB-approved protocols at the University of Pittsburgh (IRB protocol 19120198), Johns Hopkins University School of Medicine (IRB protocol 00107893), the University of Kiel, Germany (Ethikkommission der Medizinischen Fakultät der Universität Kiel, D425/07, A111/99) or obtained from LifeNet Health, an organ procurement organization (operating under the Anatomical Gift Act). All patients provided written informed consent. Liver samples for alcoholic hepatitis, alcohol-associated cirrhosis and MASLD-associated cirrhosis were obtained from liver explants in patients undergoing liver transplant. All other liver samples from living donors were obtained intraoperatively in patients in whom an intraoperative liver biopsy was indicated on clinical grounds such as during scheduled liver resection, exclusion of liver malignancy during major oncologic surgery or assessment of liver histology during bariatric surgery. The samples were frozen immediately in liquid nitrogen ensuring an ex vivo time of less than 40 s in all cases. Patients with evidence of viral hepatitis or haemochromatosis were excluded in all groups, and patients with alcohol consumption >20 g per day (for women) and >30 g per day (for men) were excluded in the normal and MASLD groups. Liver sections stained by H&E, Sirius Red and/or Trichrome were reviewed by a board-certified pathologist.
Animal studies and strains
All animal procedures were performed with approval by the Columbia University Institutional Animal Care and Use Committee (protocols, AC-AABQ5565, AC-AABQ5566), the local institutional or the Vanderbilt University Institutional Animal Care and Use Committee (protocol M2000054-01) and in accordance with the Guide for the Care and Use of Laboratory Animals; or with approval from the governmental animal care and use committees Karlsruhe, Germany (in accordance with German national guidelines on animal welfare and the regulations of the regional council Karlsruhe under permit number G-251/20). Randomization and blinding were done as described in the Reporting Summary. Mice were housed in the Irving Cancer Research Center at Columbia University and in the Central Animal Facility of the German Cancer Research Center (DKFZ) Heidelberg. They were fed with a standard mouse diet (ad libitum water and food access) with a constant temperature of 21–24 °C, 45–65% humidity and under a 12 h–12 h light–dark cycle. Lrat-cre mice have been described previously4. Clec4f-cre mice (Jax, 033296). Rspo3-floxed mice (Jax, 027313; this strain was used for crosses with Lrat-cre or Lyve1–cre), Pdgfbr-floxed mice (Jax, 010977), Rosa26–lox-stop-lox-tdTomato (TdTom) mice (Jax, 007908), Rosa26– lox-stop-lox–HBEGF (iDTR) mice (Jax, 007900), Wls-floxed mice (Jax, 012888), Lyve1-cre (Jax, 012601) and Pdgfrb-P2A-creERT2 mice (Jax, 030201) were obtained from the Jackson Laboratory. Cdh5-creERT2 mice62 as well as Mx1-cre mice, Col1a1-floxed mice, Hgf-floxed mice and Tgfbr1-floxed mice63 have been described previously. Rspo3-floxed mice (this strain was used for crosses with Pdgfrb-P2A-creERT2 mice or Cdh5-creERT2) have been described previously64. JEDI mice have been described previously19. All mouse strains, except for Wls-floxed mice (backcrossed 2× to C57BL/6) were backcrossed with C57BL/6J mice more than five times. Male mice were used between 7 and 42 weeks of age with the exception of few experiments that included female mice, as detailed in each figure legend. To induce HSC- and EC-specific deletion of Rspo3, mice were treated with 2 mg tamoxifen, dissolved in 100 μl corn oil, through oral gavage for 5 consecutive days at 8–12 weeks of age. Mice were kept for a 1-week washout period before experimental use. For the deletion of Rspo3 in hepatocytes, Rspo3-floxed mice were intravenously administrated with 1011 genomic copies per mouse of AAV8-TBG-cre (Addgene, 107787-AAV8) or AAV8-TBG-Null (Addgene, 105536-AAV8) diluted in saline solution. For the deletion of Col1a1 in the liver, Mx1-crenegCol1a1-floxed and Mx1-creposCol1a1-floxed mice received polyI:C (GE Healthcare, ww27-432-01, intraperitoneal, 10 μg g–1) three times every 3 days. Samples sizes for animal experiments differed between models and were based on estimates on the variability of specific models.
HSC depletion, liver regeneration and liver injury models
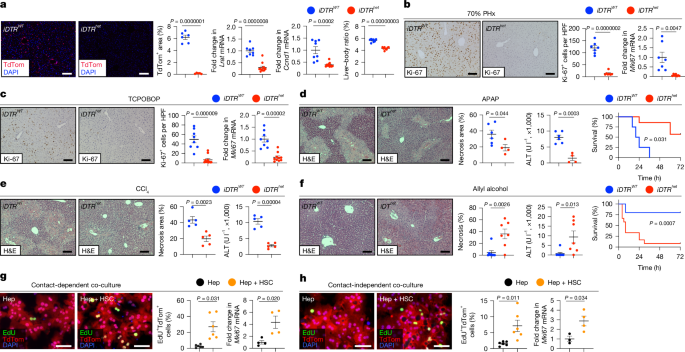
To deplete HSCs, Lrat-cre+TdTom+DTR+ mice or Lrat-cre+TdTom+DTR− littermates were injected with 0.25–2.0 ng kg−1 diphtheria toxin (DT; Sigma-Aldrich, D0564) as previously described14 and were used for experiments 7 days after DT injection unless otherwise specified. For some experiments, livers from mice with HSCs depleted using the JEDI model, described previously19, were used for analysis. For the ethanol-induced liver injury model, mice were treated with Lieber–DeCarli ‘82 ethanol-containing and control diets (Bio Serv, F1258, F1259). The ethanol diet was introduced gradually over 5 days, for a final concentration of 5%, and maintained for the indicated duration according to manufacturer’s protocol. To model metabolic dysfunction-associated steatotic liver injury, mice were fed a CDAA-HFD diet (Research Diet, A06071302) for 6 weeks. To induce MASLD-associated liver cancer, mice were fed the CDAA-HFD for 24 weeks. Tumours did not exceed the size limit of 20 mm in our IRB protocol. To model toxic liver injury, mice were treated with either CCl4 or APAP. CCl4 (Sigma-Aldrich, 319961) was dissolved in corn oil at ratio of 3:1 and injected either intraperitoneally at 0.5 ml kg−1 or given by gavage at 1.6 g kg−1. To induce severe liver fibrosis, CCl4 was dissolved in corn oil (400 µg µl−1) and delivered through oral gavage at a concentration of 1.6 g kg−1 body weight. Mice were treated twice per week for up to 20 weeks. APAP (Sigma-Aldrich, A5000) was dissolved in warm 0.9% NaCl and injected intraperitoneally into mice after overnight starvation at 300 mg kg−1 (sublethal dose) to assess liver injury or at 750 mg kg−1 (lethal dose) to determine survival. Allyl alcohol (AlOH, Sigma-Aldrich, 240532) was dissolved in 0.9% NaCl and injected intraperitoneally into mice at 60 mg kg−1 (sublethal dose) to assess liver injury or at 75 mg kg−1 (lethal dose) to determine survival. To induce liver regeneration, mice were either subjected to 70% PHx or treatment with constitutive androstane receptor ligand, 1,4-bis(2-(3,5-dichloropyridyloxy))benzene (TCPOBOP). 70% PHx was performed according to a previously published protocol65 under isoflurane and buprenorphine anaesthesia, and mice were euthanized 48 h later unless indicated otherwise. TCPOBOP (Sigma-Aldrich, T1443) was injected intraperitoneally at a dose of 3 mg kg−1 and the mice were euthanized 48 h later. For Rspo3 rescue experiments, HSC-depleted or control mice were intravenously injected with 0.5 × 1011 genomic copies per mouse of AAV8-CMV-Rspo3 (Vector Biolabs, AAV-271188) or AAV-8-CMV-EGFP (Addgene, 105530-AAV8).
Cell isolation and cell culture
Primary mouse hepatocytes were isolated as described previously14 using 15–30-week-old LSL-TdTom+ C57BL/6 mice that had been injected with 1 × 1011 GC AAV8-TBG.PI.Cre.rBG (Addgene, 107787) 7 days before isolation. Primary hepatocytes were plated and cultured in serum-free William’s E medium (Gibco, 12551-032) supplemented with hepatocyte supplement (Gibco, A13448), 10 µM dexamethasone (Gibco, A13449), 10% FBS, gentamicin and antibiotic–antimycotic (Gibco, 150062) as described previously14,66. Primary HSCs were isolated from 9–10 months old male BALB/c mice as described previously67. HSCs were seeded in 12-well plates and incubated in Dulbecco′s modified Eagle′s medium (DMEM, Gibco, 11965092) supplemented with 10% FBS, gentamicin and antibiotics. After 4 h, the medium was changed to a medium containing 0.5% FBS, followed by treated with TGFβ1 (2.5 ng ml−1, R&D systems, 240B), PDGF-BB (20 ng ml−1, R&D systems, 220BB) or IL-1β (5 ng ml−1 R&D systems, 401ML). After 24 h, cells were collected and processed for RT–qPCR. For HSC–hepatocyte co-culture, HSCs were added either directly to the tissue culture well before hepatocyte plating for contact-dependent co-culture, or into Transwell inserts (Corning, 353180) for contact-independent coculture, both at a hepatocyte:HSC ratio of 5:1. For some experiments, RSPO3 neutralizing antibody (ProteoGenix, PX-TA1446) or isotype control antibody (ProteoGenix, PTX17885) was added to the medium at a concentration of 100 nM. After 24 h of culture, EdU was added to the medium for another 24 h, followed by EdU staining according to the manufacturer’s instructions (Thermo Fisher Scientific, C10637). Images were captured using an Olympus IX71S1F-3 microscope and analysed using ImageJ software. Cells co-cultured in the absence of EdU were also collected for RT–qPCR analysis. The human HSC line LX-268 was serum-starved overnight and treated with TGFβ1 (2.5 ng ml−1, R&D systems, 240B), TGFβ2 (2.5 ng ml−1, R&D systems, 302-B2-002), TGFβ3 (2.5 ng ml−1, R&D systems, 8420-B3-005), PDGF-BB (20 ng ml−1, R&D systems, 220BB) or IL-1β (5 ng ml−1 R&D systems, 401ML) for evaluation by RT–qPCR. The mouse hepatocyte line AML12, obtained from the American Type Culture Collection, was cultured in DMEM (Thermo Fisher Scientific, 11965118) with 10% (v/v) antibiotic–antimycotic (Gibco, 150062) and 10% (v/v) fetal bovine serum (FBS; GeminBio, 900-108) at 37 °C under 5% CO2. Cell lines were regularly screened for mycoplasma contamination. Recombinant mouse RSPO3 (R&D 3500-RS) was added to the culture medium for 24–48 h at the indicated concentration. Proliferation was determined by WST-1 (Roche, 11644807001) and WNT-dependent gene expression was determined using RT–qPCR. Liver ECs were isolated from mice using a protocol similar to the HSC isolation described above but using liver perfusion medium (Gibco) and liver digestion medium (Gibco) supplemented with 20 μg ml−1 Liberase (Roche) at 3 ml min−1 for 5 min and purification of EC from the non-parenchymal cell fraction using mouse CD146 MicroBeads (Miltenyi Biotec, 130-092-007) and LS columns. Kupffer cells were purified from the non-parenchymal cell fraction after liver perfusion as described above, using magnetic mouse F4/80 MicroBeads (Miltenyi Biotec, 130-110-443) and LS columns (Miltenyi Biotec) according to the manufacturer’s instructions.
IHC, immunofluorescence and histological determination of liver fibrosis and steatosis
Liver samples were fixed with 10% formalin for paraffin-embedded blocks or with 4% paraformaldehyde for frozen blocks. Liver sections were stained with antibodies against Ki-67 (Abcam, ab16667), cyclin D1 (Abcam, ab134175), CYP1A2 (Santa Cruz, sc-53241), CYP2E1 (Abcam, ab28146), RGN (Thermo Fisher Scientific, PA5-56057), GS (Abcam, ab176562), OAT (antibodies.com, A15120), CYP2F2 (Santa Cruz, sc-374540), HAL (Sigma-Aldrich, HPA038547), E-cadherin (Cell Signaling, 3195), HNF4α (Thermo Fisher Scientific, MAI-199) or Na–K ATPase (Abcam, ab7671). Positive areas for Ki-67, cyclin D1, CYP1A2, CYP2E1, RGN, GS, OAT, CYP2F2 and HAL were analysed using ImageJ. Multiplex immunostaining was performed as previously described on 2-μm-thick formalin-fixed paraffin-embedded mouse liver sections69. The antibody elution buffer was prepared by mixing 675 μl distilled water, 125 μl 0.5 M Tris-HCl pH 6.8, 200 μl 10% (w/v) sodium dodecyl sulfate, and 8 μl 2-mercaptoethanol. The acquired images were processed and analysed using FIJI (v.2.14.0)70, ilastik (v.1.3.3post3)71 and CellProfiler (v.4.2.1)72 as described previously69. Zone-specific hepatocyte marker expression was quantified using the FIJI profile function on a portal–central vein axis, dividing the axis into nine equal sectors. Quantification was performed in four representative areas of interest for each sample. The same procedure was used to determine zonal Ki-67+ cells in Ki-67-stained liver sections, but quantifying positive cells within each sector using ImageJ. Zonal necrosis was evaluated using the above-described division of the portal–central axis into nine sectors, followed by sector-specific determination of the necrotic area assigning a percentage of necrosis (0%, 25%, 50%, 75% and 100%) to each area, based on H&E images. For determination of liver fibrosis, paraffin liver sections were stained with Picrosirius Red solution as previously described73. Frozen liver sections of 8 µm were stained in Oil Red O (Sigma-Aldrich, O9755) for 10 min. After being washed in distilled water, the sections were counterstained with Mayer’s haematoxylin for 3 min and mounted in aqueous mounting. All pictures were captured on an Olympus IX 71S1F-3 microscope coupled to a QImaging Retiga camera using QCapture Suite Plus (v.3.1.3.10) and the images were analysed using Adobe Photoshop or ImageJ software. For some analyses, images were scanned on a Leica SCN400 slide scanner with a Scanner Console (v.102.0.7.5) and quantified using ImageJ.
Determination of liver injury
Liver injury was assessed by determination of serum ALT and serum AST activity. For this, samples were measured either at the Columbia University Institute of Comparative Medicine laboratory or at the analysis centre at the University Clinic of Heidelberg. Samples were diluted with 0.9% NaCl or PBS as needed. For some experiments, the necrotic areas were determined in H&E liver sections and quantified by ImageJ software.
Immunoblotting
Proteins were extracted from liver tissue using RIPA buffer containing anti-protease (Complete, Roche) and anti-phosphatase (PhosSTOP, Roche). After adding Laemmli buffer, sonication and boiling at 95 °C, samples were loaded and run on SDS–PAGE gels and transferred onto nitrocellulose membranes (Sigma-Aldrich) using a semi-dry blotting system (Bio-Rad). The following antibodies were used: anti-ALDH2 (Proteintech, 15310-1-AP; 1:10,000), anti-RSPO3 (Proteintech, 17193-1-AP; 1:2,000), anti-GAPDH (Sigma-Aldrich, G9295; 1:75,000), anti-β-actin (Sigma-Aldrich, A3854, 1:10,000), and HRP anti-rabbit (Santa Cruz, sc-2004; 1:2,000). Blots were visualized using ultrasensitive enhanced chemiluminescent substrate (Thermo Fisher Scientific, 34094) on a FluorChem M System instrument (ProteinSimple) and quantified using FIJI.
ELISA
RSPO3 protein concentrations were quantified in homogenized mouse liver tissues using the DuoSet ELISA kit (R&D Systems). For this, lysates from snap-frozen liver samples were collected from the supernatant of homogenized tissue followed by centrifugation at 1,000g for 20 min and adjusted to 100 mg ml−1. RSPO3 protein was also determined in the supernatants from cultured primary mouse liver cells using a sandwich ELISA kit (LSBio) according to the manufacturer’s protocol. For this, supernatant samples were collected 24 h after 0.1 million cells were seeded in a 12-well plate with DMEM supplemented with 10% FCS, 1% penicillin–streptomycin and 50 mg ml−1 gentamycin, and centrifuged at 1,000g for 20 min. ELISAs were read on the iMark Microplate Reader (Bio-Rad).
Flow cytometry analysis of immune cells
Flow cytometry analysis of the lymphocytic and myeloid liver cell population was performed as previously described14,63. Briefly, liver tissues were mechanically homogenized followed by an enzymatic digestion with 1 mg ml−1 of collagenase A (Roche, 10103578001) and 0.5 µg ml−1 DNase I (Roche, 10104159001) in isolation buffer (RPMI 1640, 5% FBS, 1% l-glutamine, 1% penicillin–streptomycin and 10 mM HEPES) for 45 min at 150 rpm at 37 °C. Cells were filtered through a 100-µm cell strainer, washed and separated in two parts to analyse the myeloid and the lymphocytes cell subsets. For the latter, cells were loaded onto a Percoll gradient (67% overlay with 40%) followed by red blood cell lysis using ammonium-chloride-potassium buffer and stained. Cells were incubated with Ghost dye red 780 (Tonbo Biosciences) to exclude dead cells and anti-CD16/32 (Tonbo, 2.4G2, 1:200) before staining. The following extracellular antibodies were included: anti-CD45 (BD and BioLegend, 30-F11, 1:400), anti-CD19 (Tonbo, 1D3, 1:200), anti-CD3e (Tonbo, 145-2C11, 1:400), anti-CD4 (BD, RM4-5, 1:400), anti-CD8a (Tonbo, 53-6.7, 1:400), anti-NK1.1 (BD, PK136, 1:300), anti-CD11b (BD, M1/70, 1:500), anti-CD11c (BD, HL3, 1:200), anti-F4/80 (Tonbo, BM8.1, 1:500), anti-Ly6C (BioLegend, HK1.4, 1:500), anti-Ly6G (BioLegend, 1A8, 1:500), anti-B220 (BD, RA3-6B2, 1:200), anti-CD44 (BioLegend, IM7, 1:200), anti-CD64 (BioLegend, X54-5/7.1, 1:200), anti-CD80 (Tonbo, 16-10A1, 1:200), anti-CD86 (BD, GL1, 1:200), anti-VSIG4 (eBioscience, NLA14, 1:200) and anti-MHCII (Tonbo, M5/114.15.2, 1:400). The following intracellular antibodies were included: anti-CD3e (BD, 145-2C11, 1:400), anti-TCRβ (BD, H57-597, 1:300), anti-FOXP3 (eBioscience, FJK-16s, 1:300), anti-Ki-67 (Thermo Fisher Scientific, SolA15, 1:200) and anti-granzyme-B (BioLegend, QA16A02, 1:200). Cells were fixed using the FOXP3/transcription factor staining buffer set (Tonbo) according to the manufacturer’s protocol. The samples were analysed using the BD LSR Fortessa cell analyser. Flow cytometry analysis was performed using FlowJo (v.10.10.0).
CYP2E1 activity assay
CYP2E1 activity was analysed in liver microsomes as previously described74 with minor modifications. For microsome preparation, liver tissue was dounce homogenized in 50 mM Tris, 150 mM KCl, 2 mM EDTA buffer containing PhosSTOP (Roche, 59124500) and cOmplete Mini (Roche, 57350900). The liver homogenate was centrifuged at 6,000g for 5 min, followed by a second centrifugation of the supernatant centrifuge at 12,000g for 10 min. CaCl2 was added to the supernatant to a final concentration of 8 mM, followed by centrifugation at 211,000g for 20 min. After removal of the supernatant, the pellet was resuspended in KCl-Tris-EDTA buffer, and again centrifuged at 211,000g for 20 min. The pellet was resuspended in 0.1 ml containing 100 mM KPi, pH 7.2, 0.2 mM PNP at 37 °C for determination of CYP2E1 using the PNP method as described previously74, using a Varioskan LUX spectrophotometer (Thermo Fisher Scientific) spectrophotometer at 510 nm.
RNA isolation and RT–qPCR
Liver tissue was homogenized in a Tissuelyser (Qiagen) in Trizol and purified using chloroform, followed by isolation of total RNA using RNA isolation kits (Qiagen, Roche or Sigma-Aldrich). Total RNA from cells was isolated directly using RNA isolation kits. After quantification using a Nanodrop ND-1000 spectrophotometer, RNA was reverse-transcribed using TaqMan reverse transcription reagents (Applied Biosystems, 4368813). qPCR was run on an Applied Biosystems QuantStudio 5 Real-Time PCR system (Applied Biosystems) using PerfeCTa FastMix II buffer (Quantabio, 95120) and the following probes (Thermo Fisher Scientific): 18S (Hs99999901_s1), Acta2 (Mm001546133_m1), Aldh2 (Mm0047763_m1), Ang (Mm00833184_S1), Avpr1a (Mm00444092_m1), Axin2 (Mm00443610_m1), Ccl2 (Mm00441242_m1), Ccl3 (Mm00441259_g1), Ccl4 (Mm00443111_m1), Ccl5 (Mm01302427_m1), Ccnd1 (Mm00432360_m1), Chrna4 (Mm00516561_m1), Col1a1 (Mm00801666_g1), Col1a2 (Mm00483888_m1), Colec11 (Mm01289834_m1), Cyp1a2 (Mm00487224_m1), Cyp2e1 (Mm00487224_m1), Cyp2f2 (Mm00484087_m1), Emr1 (Mm00802530_m1), Glul (Mm00725701_s1), Gulo (Mm00626646_m1), Hal (Mm00456709_m1), Hand2 (Mm00439247_m1), Hsd3b5 (Mm00657677_mH), Il1a (Mm00439620_m1), Il1b (Mm00434228_m1), Lect2 (Mm00521920_m1), Lox (Mm00495386_m1), Mki67 (Mm01278617_m1), Oat (Mm00497544_m1), Pdgfrb (Mm00435546_m1), Rgn (Mm00485711_m1), Rspo3 (Mm00661105_m1, Mm01188251_m1), Slco1b2 (Mm00451510_m1), Tgfbr1 (Mm03024015_m1), Timp1 (Mm00441818_m1), Tnf (Mm00443258_m1), Wls (Mm00509695_m1), Wnt2 (Mm00470018_m1, Mm00437330_m1), Wnt9b (Mm00457102_m1) and RSPO3 (Hs00262176_m1).
RNA scope and spatial transcriptomics
Target RNA was detected using the RNAscope Multiplex Fluorescent Reagent Kit v2 (Advanced Cell Diagnostics (ACD), 323110) on 10 µm frozen mouse liver sections from Lrat-cre+LSL-TdTom+ mice using RNAscope Mm-Rspo3 probe (ACD, 402011) and Opal 520 Reagent (Akoya Biosciences, FP1487001KT). Anti-RFP (Rockland, 600-401-379) was used to detect TdTomato as described previously14. Confocal microscopy was performed using an AXR confocal scanner mounted on a Ti2 microscope stand (Nikon Instruments) using a ×20/0.75 Plan-Apo VC objective lens or a ×60/1.49 NA Apo-TIRF oil-immersion objective lens in the DLDRC imaging core. 100-plex spatial transcriptomics (the 100 gene panel is shown in Supplementary Table 12) focusing on WNT pathway and zonation genes was done on the Resolve platform. Probe design as well as tissue sectioning, processing, probe design and hybridization, slide imaging, spot segmentation and data preprocessing were performed as previously described29. Single-cell spatial transcriptomic analysis was performed by quantifying gene counts per cell using cell segmentation using QuPath software75. The libraries from each condition (iDTRWT, iDTRhet, Rspo3fl/fl, Rspo3ΔHSC) were integrated together using the R package Seurat76. Analysed cells included those filtered for greater than or equal to 10 gene counts per cell. After preprocessing, unbiased clustering on all 100 genes was performed using the dimensionality reduction method of principal component analysis (PCA) and uniform manifold approximation and projection (UMAP)77. Clusters corresponding to different zones were identified and annotated on the UMAP based on expression of different landmark genes as previously done29. Moreover, HSC- and EC-specific clusters were identified and annotated on the UMAP based on expression of Lrat and Pecam1, respectively. Furthermore, using the Seurat package, feature plots and violin plots were used to visualize gene expression at cluster and cell level, respectively. Lastly, once clusters were annotated, cells corresponding to specific clusters based on gene expression were mapped back onto the virtual slide to visualize spatial location of specific clusters. Expression of individual landmark genes for pericentral, midzonal and periportal zones were used to verify accurate spatial localization of clusters. For zone- and cell-specific analysis of Rspo3 expression, the overall expression of established landmark genes was first used to define zones, followed by assignment of cells to one of three zones based on their localization within these regions. This was done by plotting cells positive for Cyp2f2 on one image using ggplot2 in R, and cells positive for Cyp2e1 on another ggplot2 image. These images were processed in MATLAB using the Image Processing Toolbox to smooth and fill in distinct regions as either zone 1 or zone 3, yielding two matrices delineating zone 1 and zone 3 regions. Cells were classified as zone 1 if they were located in the zone 1 region only, zone 3 if they were located in the zone 3 region only, and zone 2 if they were located in overlapping zone 1 and 3 regions. Gene expression levels, including Rspo3, were evaluated across both zone and cluster using the VlnPlot function in Seurat.
Microarray and bulk RNA-seq, heat maps and pathway analysis
RNA-seq analyses were performed on high-quality total RNA samples, with RNA integrity numbers of >8 (determined using a Bioanalyzer 2100, Agilent Technologies). Bulk RNA-seq data were processed by the Columbia Genome Center. For each sample, a minimum of 20 million 100 bp single-end reads were sequenced on the Illumina NovaSeq 6000 system. RTA (Illumina) was used for base calling and bcl2fastq2 (v.2.19 and v.2.20) was used for converting BCL to fastq format, coupled with adaptor trimming. A pseudoalignment to a kallisto index was created from transcriptomes (human, GRCh38; mouse, GRCm38) using kallisto (v.0.44.0). To explore similarities and dissimilarities between samples, count data were normalized using the variance stabilizing transformation function from the DESeq2 package. Microarray analysis for comparison between Ctnnb1ΔHep and Ctnnb1fl/fl livers was conducted using a published dataset (GSE68779). Heat maps were generated using the Heatmapper tool78. KEGG pathway analysis in HSC-depleted liver was performed using enrichr using the 100 significant (P < 0.05) genes with the highest combined log-transformed fold change in HSC iDTR- and JEDI-depleted livers compared with their respective controls (GSE211370)79.
GSEA
GSEA was performed using GSEA v.4.3.2 software (https://www.gsea-msigdb.org/gsea/downloads.jsp). Analysis was performed from pre-ranked genes from DESeq2 (v.1.42.0) analysis of bulk RNA-seq data using the GSEAPreranked function with 1,000 permutations on the Hallmark collection from the Molecular Signature Database (MSigDB) or by curating the CTNNB1_Liver gene set from genes with a log-transformed fold change of >1 in the microarray analysis from Ctnnb1ΔHep and Ctnnb1fl/fl livers as reference for β-catenin-regulated liver genes.
Nucleus isolation for snRNA-seq
For snRNA-seq analysis, human and mouse livers (Supplementary Tables 4 and 10) were processed as previously described14. In brief, frozen liver tissue was minced with scissors in 1 ml TST buffer in the well of a six-well plate for 10 min on ice. The homogenized solution was then passed through a 40-μm cell strainer. An additional 1 ml of TST buffer and 3 ml of 1× ST buffer were used to wash the well and passed through a 40-μm cell strainer. The resulting 5 ml of nuclei suspension was centrifuged for 5 min at 500g at 4 °C. The supernatant was discarded, and the pellet resuspended in 1 ml of 1× ST buffer. The nuclei suspension was then passed through a 35 μm filter. For single-cell multiome ATAC plus gene expression analysis, the Chromium Nuclei Isolation with RNase Inhibitor Kit (10x Genomics, PN-1000494) was used to isolate nuclei from mouse liver.
scRNA-seq, snRNA-seq and scATAC and gene expression sequencing
All analysed scRNA-seq data, including HSCs isolated from 1× CCl4-, 2× CCl4-, 4× CCl4– and 12× CCl4-treated mouse livers (GSE172492) and 0, 15, 30 and 34 weeks of HF-HFD-treated mouse livers (GSE166504) as well as whole mouse liver (GSE158183) have been published and deposited previously. Samples for human snRNA-seq analysis (GSE256398) were prepared as previously described14 using the 10x Chromium Single Cell Platform using a ChromiumSingle Cell 3′ Library and Gel Bead Kit v.3 and a Chromium Single Cell B Chip kit (10x Genomics, PN-1000074) according to the manufacturer’s protocol. Data were aligned to a modified version of the GRCh38 reference genome (counting intronic reads as well as those aligned to exons), and estimated cell-containing partitions and associated unique molecular identifiers (UMIs) using Cell Ranger v.3.1.0 from 10x Genomics.
snRNA-seq and scRNA-seq analysis
In total, 26 human snRNA-seq datasets and 4 mouse snRNA-seq datasets were analysed (Supplementary Table 10). Technical artefacts such as ambient background RNA and empty droplets in these datasets were removed using the remove-background function in CellBender v.0.2.0 as described (fpr = 0.01 for human datasets; fpr = 0.1 for mouse datasets)14. For human datasets, the output raw_feature_bc_matrix_filtered.h5 from CellBender for each sample was further subjected to doublet removal using Scrublet with the default parameters80. The resulting singlets from human datasets and the raw_feature_bc_matrix_filtered.h5 from mouse datasets were analysed in Seurat (v.5.0.1).
For each dataset, the samples were combined and low-quality cells or outlier cells were filtered using the same standard (nFeatureRNA < 200 or nFeatureRNA > 6500 or nCount_RNA > 40000 or percent.mt > 20 for human, nFeatureRNA < 200 or nFeatureRNA > 7500 or nCount_RNA > 60000 or percent.mt > 20 for mouse). Each sample was first analysed in parallel using the NormalizeData, FindVariableFeatures (nFeatures = 3000), ScaleData and RunPCA function. The samples were integrated using the SelectIntegrationFeatures, FindIntegrationAnchors (k.anchor = 10, reduction = “rpca”) and IntegrateData functions. After integration, a single integrated analysis with a batch-corrected integrated count matrix layer was run on all cells using the ScaleData, RunPCA (npcs = 50), RunUMAP, FindNeighbors and FindClusters functions. All of the default parameters were used unless mentioned otherwise. The main cell types including T cell/natural killer, myeloid cells, HSCs, ECs, cholangiocytes and hepatocytes were identified manually by checking the expression of well-known marker genes as described previously14. For each main cell type, the clusters were subset and reclustered using the non-batch-corrected RNA count matrix layer using ScaleData, RunPCA (npcs = 50), RunUMAP, FindNeighbors and FindClusters function. Detailed cell types were identified manually using the expression of well-known marker genes as described previously81 and markers for each population are provided in the Supplementary Information as dot plots for mouse snRNA-seq data (Supplementary Information 4) and human snRNA-seq data (Supplementary Information 5). Clusters that simultaneously express marker gene sets from two or more cell types were identified and further removed as doublets. For each dataset, the differentially expressed genes (DEGs) in every cell cluster were identified using the FindAllMarkers function in Seurat v.3.
Some published datasets, including human and mouse snRNA-seq from the Henderson lab82 and from the Livercellatlas81 were analysed using web-based tools described in the respective publications.
CellPhoneDB analysis
After identifying cell types in each dataset as described above, we used CellPhoneDB83 in the most recent v.5 version to identify ligand–receptor interactions in n = 6 healthy livers from our human snRNA-seq dataset (GSE256398). To determine mouse ligand–receptor interactions, mouse genes from n = 2 healthy control livers (GSE256398) were first converted to human gene symbols (HGNC) using biomaRt (v.2.60.1) package in R, followed by recommended procedures for preparation of input files. All CellPhoneDB statistical analysis was performed using the default parameters and the percentage cell expression threshold of 5%. After ranking cell–cell interactions by the interaction scores, all interactions with a positive interaction score were further ranked by the mean expression. Heat maps showing ligand–receptor interactions, log2-transformed mean (molecule 1, molecule 2) and log10[P] values were generated using ggplot2 (v.3.4.4) package.
Gene expression and survival analysis in clinical cohorts of patients with CLDs
To determine survival and liver-related events in the SteatoSITE cohort of patients with MASLD84, normalized counts per minute of RSPO3 from the biopsy subset of cases in the SteatoSITE data commons were used in survival analysis using R (v.4.3.0) in RStudio (v.2023.12.0 build 369) and the ‘survminer’ package (v.0.4.9). The optimal cutpoint for normalized RSPO3 counts was separately calculated for overall survival, and hepatic decompensation (when a first coding of any component of the composite outcome occurred after the biopsy date and with death as a competing risk) using surv_cutpoint, applying maximally selected rank statistics of the ‘maxstat’ package (v.0.7-25) with a minimum proportion of 0.25. Kaplan–Meier estimator curves of all-cause mortality were compared by regular log-rank testing with weights = 1. To determine survival in the dbGaP phs001807.v1.p1 cohort of patients with ALD85, the median RSPO3 expression was established as a threshold for high and low expression cohorts, for analysis of survival using Kaplan–Meier estimator curves of all-cause mortality and log-rank testing. Moreover, genome-wide hepatic transcriptome datasets of clinical cohorts of MASLD without HCC (Gene Expression Omnibus (GEO): GSE49541 (ref. 86) and GSE193066 (ref. 87)) and with resected or ablated HCC (GSE192959 (ref. 87)), alcoholic-associated cirrhosis (GSE103580 (ref. 88)) and severe alcohol-associated hepatitis (GSE94397 (ref. 88)) were analysed for association with clinical disease phenotypes and outcome for RSPO1, RSPO2, RSPO3 and RSPO4 genes, including association with known WNT pathway target genes (CYP2E1 and CYP1A2), and previously reported transcriptomic signatures of overall survival, decompensation and HCC risk in CLDs (prognostic liver signature (PLS))89 and overall survival in severe alcohol-associated hepatitis88. For analyses in these cohorts, high expression of the RSPO genes was defined based on a top-quartile cut-off in each cohort; the presence of high-risk pattern of the prognostic transcriptomic signatures was determined by the nearest template prediction algorithm90; associations of the high gene expression and presence of the high-risk signatures with clinical phenotypes and outcomes were evaluated using Wilcoxon rank-sum tests, log-rank tests and/or the Kaplan–Meier method depending on the availability of clinical annotations in each cohort.
Genome-scale metabolic pathway analysis
We performed metabolic pathway analysis using liver transcriptomics data, comparing JEDI HSC-depleted livers to their controls (GSE211370), hepatocytes from Rspo3fl/fl and Rspo3ΔHSC mouse livers (GSE256398) and livers from Ctnnb1ΔHep and Ctnnb1fl/fl mice (GSE68779), as described previously91. In brief, we translated the statistically significant DEGs (FDR ≤ 0.05) to enzymatic reaction rate changes according to gene–protein–reaction (GPR) rules. As a database for pathways, reactions and GPR rules, we used the Mouse1 (v.1.3.0) metabolic models, genome-scale metabolic reconstruction92. Metabolic pathways were then scored and ranked according to the amount of perturbed reactions that they encompass91. We also computed P values for each pathway to evaluate their statistical significance. We used the hypergeometric test, which is based on the hypergeometric distribution91. The computed P values were subjected to a FDR correction, using the Benjamini–Hochberg procedure.
Bulk and spatial metabolomics
Snap-frozen mouse liver samples were analysed at the Roswell Park Comprehensive Cancer Center Bioanalytics, Metabolomics and Pharmacokinetics Shared Resource, using the MxP Quant 500 XL kit (Biocrates Life Sciences) according to the manufacturer’s instructions. In brief, liver samples were homogenized at a ratio of 1 mg of tissue to 3 μl of solvent (25% ethanol and 75% 0.01 M phosphate buffer) using optimized settings on the Omni-Bead Ruptor 24 (Omni). After centrifugation, 10 μl of each supernatant, quality control samples, blank, zero sample or calibration standard were added on the filterspot (already containing internal standard) in the appropriate wells of two 96-well plates and dried under a gentle stream of nitrogen. On one plate, the samples were derivatised with phenyl isothiocyanate for the amino acids and biogenic amines, and dried again. Sample elution on both plates was performed with 5 mM ammonium acetate in methanol. Sample extracts were diluted with either water for the HPLC–MS/MS analysis (1:1) or kit running solvent (Biocrates Life Sciences) for flow injection analysis (FIA)–MS/MS (50:1), using the Shimadzu HPLC system interfaced with the Sciex 5500 mass spectrometer. Data were processed using WebIDQ software (Biocrates Life Sciences), and Limma for differential metabolite analysis.
For DESI MS imaging, fresh-frozen mouse liver tissue blocks were embedded in 5% gelatin over dry ice and stored at −80 °C. The tissue blocks were then cryosectioned at a thickness of 8 µm (Leica, CM3050S) and thaw-mounted onto a microscope slide and stored at −80 °C until analysis. The microscope slides were dried in a vacuum desiccator for 8 min. DESI MSI data acquisition was performed on the Synapt G2-XS QToF mass spectrometer coupled to a DESI ion source (Waters) in positive-ion sensitivity mode with a mass range of m/z 100–1,000. The following DESI parameters were used: capillary voltage and sampling cone voltage of 0.5 kV and 50 V, respectively, DESI sprayer angle of 78 °C, source temperature of 150 °C, nebulizing gas (N2) pressure of 0.9 bar and spatial resolution of 40 µm. The solvent used was methanol:water 95:5 (v/v) with 0.01% formic acid and 40 pg µl−1 leucine enkephalin, at a flow rate of 1.5 µl min−1. Data were processed and visualized in HDI imaging software (Waters, v.1.6) and SCiLS (Bruker, version 2024a). Peak picking and lockmass correction were performed using leucine enkephalin ([M+H]+, m/z 556.2771). Lipid annotations were performed by accurate mass search against the Lipidmaps database93 and also from annotations previously performed by lipidomics analysis using UPLC with ion mobility ToF MSE(HDMSE) data-independent acquisition94. Co-localizing with glutamine synthetase by IHC was used to determine zonation in images. The distribution of the intensity of the ions and quantification using area under the curve of the corresponding lipid feature, denoting the average intensity of the m/z interval within the tissue, normalized to TIC, were generated within the SCiLS software.
Quantification and statistical analysis
No statistical methods were used to predetermine sample size. Investigators were blinded for in vivo treatments and post-mortem analyses such as (1) quantification by IHC and (2) determination of gene expression by qPCR. Investigators were not blinded for snRNA-seq analyses studies as there were not separate groups involved or the samples were annotated. For immunoblotting, the investigators were not blinded when loading the gel to display the results in a logical way. Statistical significance was determined using GraphPad Prism (v.9.0) or R (v.4.0.2). After assessing the normal distribution of the data using D’Agostino and Pearson omnibus normality tests and or Shapiro–Wilk’s normality test, P values were calculated, and all statistical tests used are described in the figure legends. Survival curves were represented using the Kaplan–Meier method and compared using log-rank statistics.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.