Chemicals
Compounds were obtained from the following vendors: Sigma-Aldrich: oxymorphone (O-004-1ML), loperamide (1448005), methadone hydrochloride (M0267), morphine sulfate pentahydrate (M8777), sufentanil citrate (SML0535), herkinorin (5.08018.0001), buprenorphine (B9275), serotonin hydrochloride (H9523), carbachol (C4382), naloxone hydrochloride (N7758) and GDP (G7127). Cayman Chemical: PZM21 (20576-10), fentanyl citrate (22659) and GTPγS (35098). DAMGO (11711) was from Tocris Bioscience. Oliceridine (TRV-130; 510256) was from MedKoo Biosciences. Dynorphin A 1-17 (3195) was from Fisher Scientific. Somatostatin-14 was custom synthesized by CPC Scientific. Met-enkephalin (30-0-10) was from American Peptide Company. SR-17018, muzepan1 and muzepan2 were made in house. 35S-GTPγS (NEG030H001MC) was from Revvity. 3H-(-)naloxone was provided by the National Institute on Drug Abuse (NIDA) Drug Supply Program.
Animals
Male and female C57BL6/J (JAX:000664) and male MOR-KO (JAX:007559) mice were purchased from The Jackson Laboratory. MOR-KO mice were maintained by homozygous breeding. Mice were housed in groups of 2–5 and maintained on a 12 h:12 h light:dark cycle with food and water ad libitum. All adult mice were naive and at least ten weeks old prior to injection. For thermal antinociception tests, investigators were blinded to drugs and doses being administered. Mice were administered drugs intraperitoneally at a volume of 10 µl per g; for combinations of drugs, a single solution was prepared. All mice were used in accordance with the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals with approval by The Herbert Wertheim UF Scripps Institute of Biomedical Technology and Innovation Animal Care and Use Committee.
Cell lines
Chinese Hamster Ovary (CHO-K1) cells were purchased from ATCC. Human MOR (hMOR), mouse MOR (mMOR) and human KOR (hKOR) cells have been described previously24. For the other cell lines, receptor constructs were purchased from cDNA Resource Center including SST2R (SSTR20TN00), 5-HT1AR (5TR01ATN00) and M2R (MAR020TN00). The SSTR2R, 5-HT1AR and M2R cell lines were produced by electroporation of a pcDNA3.1 vector containing the N-terminally HA-tagged receptor into the parental cell line. A BD FACSAria3 flow cytometer was utilized to select for individual cells expressing receptor utilizing an anti-HA AlexaFluor 488 conjugate antibody (1:200). hMOR, hKOR, SST2R, 5-HT1AR and M2R cell lines were maintained under geneticin selection (500 µg µl−1). The mMOR cell line was maintained under puromycin selection (500 µg µl−1). Cells were maintained in 1:1 DMEM:F12 media supplemented with 10% heat-inactivated fetal bovine serum (HI-FBS) at 37 °C with 5% CO2. Prior to use in assays, cells were serum-starved for 30 min (hMOR, hKOR, 5-HT1AR and M2R) or 2 h (mMOR and SST2R) then removed from the plate with 5 mM EDTA in PBS with a scraper. Pellets were rinsed with PBS and frozen in 1.5 ml tubes at −80 °C until use. All cell lines were verified to be mycoplasma-free by monthly testing.
35S-GTPγS binding and release
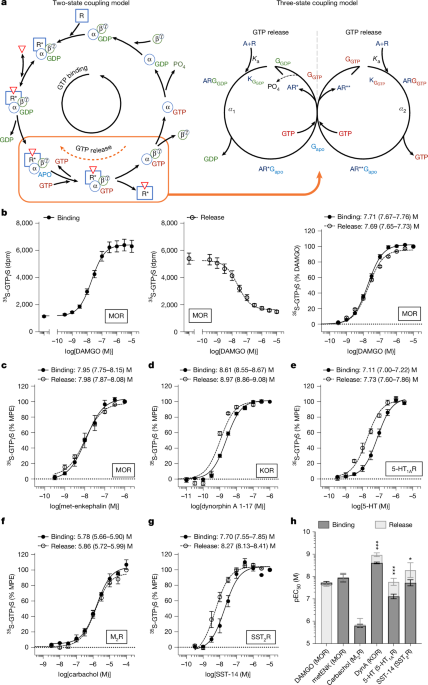
35S-GTPγS binding
35S-GTPγS binding in cell lines was performed similarly as previously described16,24. In brief, for CHO-hMOR cells, pellets were homogenized with a Potter–Elvehjem Teflon-on-glass Dounce homogenizer in homogenization buffer (10 mM Tris (pH 7.4), 100 mM NaCl, 1 mM EDTA) then pelleted (20,000g, 4 °C, 30 min). All other cell lines (CHO-mMOR, CHO-hKOR, CHO-hSST2R, CHO-h5-HT1AR and CHO-hM2R) were homogenized in 10 mM Tris (pH 7.4), 1 mM EDTA. All reactions were performed with 0.1% DMSO and 0.1 nM 35S-GTPγS (specific acitivity = 1,250 Ci mmol−1; Revvity) in 50 mM Tris (pH 7.4), 100 mM NaCl, 5 mM MgCl2, 1 mM EDTA with differing quantities of protein and GDP. For CHO-hMOR, CHO-mMOR and CHO-5-HT1AR cells, 10 µg protein and 10 µM GDP was used. CHO-M2R cell reactions were performed with 3 µg protein and 3 µM GDP. CHO-SST2R cell reactions were performed with 10 µg protein and 20 µM GDP. CHO-hKOR cell reactions were performed with 15 µg protein and 3 µM GDP. Reactions were terminated by rapid filtration through GF/B filters with cold water after a 1 h incubation. Filters were punched into 96-well Opti-plates (Revvity) and dried overnight. Radioactivity was measured with 100 µl per well MicroScint-20 on a MicroBeta 2 (Revvity). For determination of bias between GTPγS and β-arrestin2 recruitment, 35S-GTPγS binding was performed in CHO-hMOR cells exactly as described24 and is shown in Extended Data Fig. 3a.
For 35S-GTPγS binding in C57BL/6J and MOR-KO spinal cord, tissue was homogenized via a Polytronic Tissue Tearor (BioSpec Products, 985370) and then a glass-on-glass Dounce homogenizer in homogenization buffer (10 mM Tris (pH 7.4), 1 mM EDTA). Homogenate was pulled through a 28G insulin needle before pelleting at 20,000g, 4 °C, 30 min. Reactions were performed with 10 µg, 10 µM GDP, 0.1% DMSO and 0.1 nM 35S-GTPγS and incubated for 1 h at 25 °C. Reactions were terminated as described above.
35S-GTPγS release in sodium-free conditions
Membranes were prepared in 10 mM Tris (pH 7.4), 1 mM EDTA as described for 35S-GTPγS binding in (10 mM Tris (pH 7.4), 1 mM EDTA). For 35S-GTPγS loading (the ‘pulse’) of CHO-hMOR, CHO-mMOR and CHO-5-HT1AR cells, 1 mg of protein was incubated with 1 nM 35S-GTPγS and 10 µM GDP in 20 ml of 50 mM Tris (pH 7.4), 5 mM MgCl2, 1 mM EDTA for 1 h at 25 °C. Identical conditions were used for CHO-SST2R cells except GDP was increased to 20 µM. For CHO-M2R cells, 0.3 mg of protein was incubated in the same conditions with 3 µM GDP. For CHO-hKOR cells, 1.5 mg of protein was incubated with 3 µM GDP. Release was performed by diluting tenfold into 50 mM Tris (pH 7.4), 100 mM NaCl, 5 mM MgCl2, 1 mM EDTA supplemented with 1 µM cold GTPγS and GDP corresponding to the receptor utilized. Release was performed for 1 h at 25 °C then terminated as described for 35S-GTPγS binding. See Supplementary Fig. 1 for a schematic.
35S-GTPγS release following 100 nM DAMGO-stimulated loading
For CHO-mMOR cells and C57BL/6J spinal cord studies relying on 100 nM DAMGO for loading, membranes were prepared as described for the sodium-free loading conditions. Then, 1 mg of protein was incubated with 1 nM 35S-GTPγS, 10 µM GDP, and 100 nM DAMGO in 2 ml of 50 mM Tris (pH 7.4), 100 mM NaCl, 5 mM MgCl2, 1 mM EDTA for 1 h at 25 °C. Release was performed in large-volume 96-well plates with 10 µg protein, 10 µM GDP, 0.1% DMSO, at 2 ml final volume and incubated for 1 h at 25 °C. Reactions were terminated as described above for 35S-GTPγS binding. See Supplementary Fig. 2 for a schematic.
Radioligand binding
3H-(-)naloxone binding studies were performed as previously described16. Membranes were prepared via homogenization with a Polytronic Tissue Tearor then glass-on-glass Dounce homogenization in homogenization buffer (50 mM Tris (pH 7.4), 1 mM EDTA). Homogenate was pulled through a 28G insulin needle before pelleting at 20,000g, 4 °C, 30 min. Binding was performed with 10 µg membrane in 10 mM Tris (pH 7.4) containing 1% DMSO and approximately 2 nM 3H-naloxone (1.50–2.04 nM; specific acitivity = 48.19 Ci mmol−1) at a final volume of 200 µl. Reactions were incubated for 1 h at 25 °C then filtered through GF/B fiberglass filters with cold 10 mM Tris (pH 7.4) by rapid filtration over GF/B filters and washed with cold 10 mM Tris buffer. Filters were punched into white, 96-well OptiPlate and dried overnight. Radioactivity was quantified using 100 µl per well MicroScint-20 on a MicroBeta2. The Kd of 3H-(-)naloxone determined by homologous competition in these studies is 0.83 (0.33–1.1) nM, n = 6.
β-arrestin2 recruitment
β-arrestin2 recruitment was performed as previously described24. In brief, U2OS-β-arrestin2-hMOR PathHunter cells were plated at a density of 5,000 cells per well in a 384-well, white-walled assay plate in OptiMEM supplemented with 1% HI-FBS and incubated at 37 °C with 5% CO2 for 16–20 h. Drug was prepared in PBS and cells were treated for 90 min at 37 °C. β-arrestin2 recruitment was determined using the PathHunter Detection Kit and luminescence was measured using a BioTek Synergy Neo2 multimode plate reader (BioTek).
Cytochrome P450 inhibition
Inhibition studies were carried out with 10 µM compound incubated with human liver microsomes and selective marker substrates (1A2, phenacetin demethylation to acetaminophen; 2C9, tolbutamide hydroxylation to hydroxytolbutamide; 2D6, bufuralol hydroxylation to 4′-hydroxybufuralol; 3A4, midazolam hydroxylation to 1′-hydroxymidazolam). After a 10 min incubation, the reaction was terminated and the percent inhibition was determined as previously described34.
Antinociception
Thermal antinociception was performed as previously described24. Prior to testing, mice were habituated to the testing room for 1 h. The tail flick test was determined as the amount of time until a mouse rapidly flicked its tail when placed 2–3 cm into a 49 °C water bath with a cut-off applied at 30 s. The hot plate test was measured using a 52 °C hot plate analgesia meter (Columbus Instruments) and forepaw or hindpaw licking or flicking were observed with a maximum latency of 20 s to prevent tissue damage.
Respiration and heart rate measures
Respiratory and heart rate parameters were simultaneously measured using the MouseOx Plus pulse oximeter (Starr Life Sciences) as previously described16,24. Two days prior to testing, mice were shaved around the neck and habituated for 30 min to the collars and 50 ml conical tubes which were modified to restrain the mice. The following day, the mice were habituated to the collars and restraint for 30 min. On testing day, the basal vital signs of the mice were determined for 30 min then animals were injected and monitored for 90 min.
Pharmacokinetics
Male C57BL6/J mice were injected intraperitoneally with muzepan1 or muzepan2 at the doses indicated and blood was collected at indicated time points. Brains were collected following cervical dislocation, and snap frozen in liquid nitrogen. Samples were subjected to liquid chromatography (Shimadzu)–tandem mass spectrometry from AB Sciex. Pharmacokinetic parameters were calculated using a noncompartmental model24 (Phoenix WinNonlin, Pharsight).
Data analysis
Concentration response studies were analysed by nonlinear regression analysis following normalization (baseline = 0 and maximum response = 100%). For all of the studies, we used the mean of the individual experiments to generate the potency (logEC50) and efficacy (Emax) values as presented as pEC50 with 95% confidence interval in the figures and graphs and as pEC50 with s.e.m. in the table. Both potency and efficacy parameters were produced using three-parameter nonlinear regression with adaptation of the equation:
$$\mathrm{Response}=\mathrm{basal}+\frac{{E}_{\max }-\mathrm{basal}}{{10}^{({\mathrm{logEC}}_{50}-X)}+1}$$
where X is the agonist concentration in log molar units and logEC50 is agonist potency in log molar units. Statistical analyses comparing binding and release parameters, for each compound, was performed by unpaired t-test. For the MOR studies, DAMGO was tested in parallel for all compounds and was used for normalization (baseline = 0, DAMGO at 10 µM = 100%). Statistical comparisons between binding and release parameters were performed by unpaired t-test comparing the individual parameters determined in each experiment; the number of replicates are indicated in the table.
In addition, a form of the operational model frequently applied to bias analysis was employed as the binding and release assays were considered independent measures of agonist activity17,35. The equation takes the form:
$$\mathrm{Response}=\mathrm{basal}+\frac{{E}_{\max }-\mathrm{basal}}{1+{\left(\frac{1+{10}^{(X+\log K)}}{{10}^{(X+\log {R}_{\text{reference}}+\Delta \log R)}}\right)}^{n}}$$
where basal and Emax describe the system limits, and n defines the transducer slope. For the reference agonist DAMGO, logK and ΔlogR are held constant at zero. In this case, the logRreference for the reference agonist reduces to the plogEC50. For full test agonists the logRreference is held constant, from the fit of the reference agonist, and the ΔlogR is permitted to float. The logK is held constant at zero for all full agonists. For partial test agonists, the logRreference is again held constant and the ΔlogR and logK are permitted to float.
For the determination of the transduction efficiency, the ΔlogR was determined for each individual assay with DAMGO serving as the reference agonist. The ΔΔlogR was determined by unpaired t-test between the ΔlogR from the G-protein release assay and the ΔlogR of the G-protein binding assay. The same approach was used to determine the bias factor comparing the ΔlogR in G-protein binding versus the ΔlogR in β-arrestin2 recruitment in the CHO-hMOR cells.
In radioligand binding studies, naloxone competition was fit to the homologous (naloxone) or heterologous (muzepan1, muzepan2) competition equation:
$$\mathrm{Binding}=\mathrm{bottom}+\frac{{B}_{\max }\times [{}^{3}\text{H-naloxone}]}{[{}^{3}\text{H-naloxone}]+{10}^{\log (X)}+{K}_{\mathrm{naloxone}}}$$
where, for 3H-naloxone binding, ‘bottom’ and Bmax are the non-specific and maximum binding, [3H-naloxone] is the radioligand concentration, Knaloxone is the naloxone equilibrium dissociation constant, and X is the cold naloxone concentration in molar units. For muzepan1 and muzepan2, competition data were fit to the heterologous competition equation:
$$\mathrm{Binding}=\mathrm{bottom}+\frac{\mathrm{Top}\,-\,\mathrm{bottom}}{1+\frac{{10}^{\log (X)}}{{10}^{{K}_{i}\times \left(1+\frac{[{}^{3}{\rm{H}}-\mathrm{naloxone}]}{{K}_{\mathrm{naloxone}}}\right)}}}$$
where parameter definitions are shared between the two equations. In the heterologous competition equation, Top is the maximum observed binding, Ki is the molar affinity constant of the competitive ligand, and X is the concentration of the competitive ligand. Experiments were run together and both [3H-naloxone] and Knaloxone are held constant for the analysis.
Antinociception
A maximum possible effect (%MPE) was calculated as 100% × [(baseline response − test response)/(cut-off time − baseline response)]. For the determination of potency, the %MPE was compared at the 1 h time point by nonlinear regression analysis in GraphPad Prism (v.10.4), sharing the Hill slope and constraining the bottom to 0 and the top to 100%. For the comparison of morphine potency with and without muzepan1, the AUC was determined from the %MPE over the 4 h testing period; this was normalized to the maximum possible effect (the AUC if all points reached 100%) and fit the nonlinear regression analysis. Statistical comparisons of the logED50 were made between two curves in Prism using an extra-sum-of-squares F test.
Respiration and heart rate
A two-way repeated measures ANOVA was used to compare drug effects (35–120 min) as a function of time and the results are presented in Extended Data Table 5. In addition, the AUC was determined by normalizing to the first 30 min of habituation for the following drug effect over 1 h. These values were then compared by one-way ANOVA comparing to vehicle with a Dunnett’s post hoc test, or between drug treatments (fentanyl versus fentanyl plus 3 mg kg−1 or 24 mg kg−1 muzepan1; 3 groups, Tukey’s post hoc test).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.