Animals
Aged C57BL/6 mice (16–21 months old) were obtained from the National Institute on Aging rodent colony. Young C57BL/6 mice (3 months old) were obtained from Jackson Laboratories. All experiments used male mice. All mice were kept on a 12 h–12 h light–dark cycle and provided ad libitum access to food and water. All animal care and procedures complied with the Animal Welfare Act and were in accordance with institutional guidelines and approved by the Veterans Affairs Palo Alto Committee on Animal Research and the institutional administrative panel of laboratory animal care at Stanford University.
Human tissue
Post mortem fresh-frozen brain tissues were obtained from Stanford/VA Aging Clinical Research Center with approval from the Stanford Institutional Review Board and patient consent. Autopsies were performed no more than 12 h after death, and all samples used in this study were stored at −80 °C until the time of processing. Group characteristics are summarized in Supplementary Data 2. Individuals in the Alzheimer’s disease group were both clinically diagnosed and pathologically determined to exhibit Alzheimer’s disease brain hallmarks including β-amyloid and tau pathophysiology.
Transmission electron microscopy
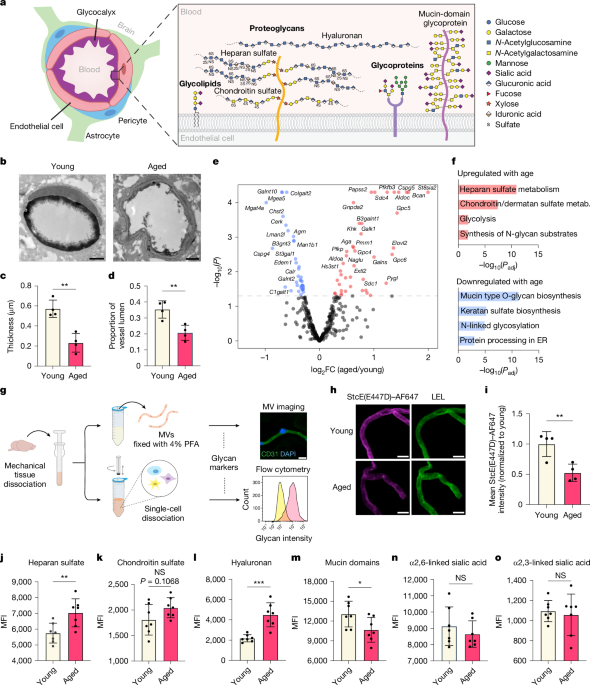
Transmission electron micrographs of the brain endothelial glycocalyx layer were obtained as described4 with some modifications. In brief, young (3-month-old) and aged (21-month-old) mice were perfused with ice-cold fixation buffer composed of 2% glutaraldehyde (EMS), 2% sucrose, 0.1 M sodium cacodylate buffer (EMS), and 2% lanthanum nitrate (EMS) through the left ventricle using a peristaltic pump at 2 ml min−1. The brain was removed and sliced coronally using a matrix into 1-mm-thick sections. Cortical punches (1 mm3) were cut and immersed in perfusion solution for 2 h before storage overnight in perfusion solution without glutaraldehyde at 0 °C. The following day, the samples were washed in a 0.03 M NaOH 2% sucrose solution and subjected to an ascending ethanol gradient and embedded in epoxy resin (EMS). 90 nm sections were cut using a Leica UC6 ultramicrotome (Leica Microsystems) and collected onto formvar-coated 50-mesh copper grids. The grids were post-stained with 2% uranyl acetate followed by Reynold’s lead citrate for 5 min each. Sections were imaged using a Tecnai 12 120 kV TEM (FEI), and data were recorded using either an UltraScan 1000 with Digital Micrograph 3 software (Gatan) or a Rio16 CMOS camera with GWS software (Gatan). Between 6 and 12 cortical capillaries per animal were captured by a blinded observer. Quantitative analysis of endothelial glycocalyx thickness and area were performed using ImageJ software (Extended Data Fig. 1a). For StcE treatment experiments, 3-month-old mice were retro-orbitally injected with 0.25 mg kg−1 StcE or inactivated StcE(E447D) 24 h before perfusion.
For BBB ultrastructural analysis, mice were injected retro-orbitally with 0.3 ml of 0.5 mg g−1 of HRP type II in PBS (Sigma, P8250). After 30 min, brains were dissected and fixed in 0.1 M sodium cacodylate buffer (EMS) with 5% glutaraldehyde (EMS) and 4% PFA (EMS) for 1 h at room temperature and then for 16 h in 4% PFA/0.1 M cacodylate at 4 °C. Tissues were washed overnight with 0.1 M sodium cacodylate at 4 °C and then sliced coronally using a matrix into 1-mm-thick sections. Cortical punches (0.5–1 mm3) were cut and incubated in 0.5 mg ml−1 of 3,3′-diaminobenzidine with 0.01% hydrogen peroxide in TBS for 45 min at room temperature. Tissues were washed with TBS overnight, post-fixed in 2% osmium tetroxide and 2.5% potassium ferrocyanide in 0.1 M sodium cacodylate, and en bloc stained with 1% uranyl acetate and Walton’s lead aspartate stain. Samples were then dehydrated in an ascending ethanol gradient and embedded in epoxy resin (EMS). Eighty-nanometre sections were cut using a Leica UC7 ultramicrotome (Leica Microsystems) and collected on formvar-coated 100-mesh copper grids. The grids were post-stained with 3.5% uranyl acetate followed by Sato’s lead citrate. Sections were imaged using a Tecnai 12 120 kV TEM (FEI), and data were recorded using a Rio16 CMOS camera with GWS software (Gatan). The assessment of tight junctions in the images was performed in a blinded manner.
Flow cytometry on acutely isolated brain endothelial cells
Microvessels from young (3-month-old) and aged (21-month-old) mice were isolated as previously described with some modifications26,34. In brief, mice were euthanized via CO2, and brains were retrieved in PBS supplemented with 1% bovine serum albumin (BSA) and 1× cOmplete protease inhibitor cocktail (Millipore Sigma) on ice. The olfactory bulb was discarded, and meningeal vessels were removed by gentle rolling on blotting paper. Brains were minced using a razor blade on ice and then homogenized using a loose-fit, 7 ml Dounce (Wheaton) in 1% BSA-PBS with 1× protease inhibitor. The homogenate was centrifuged in 27% (wt/vol) 70 kDa dextran (Sigma) in HBSS at 4,400g for 25 min. Myelin and parenchymal cell layers were removed. Pelleted microvessels were deposited on a pre-wet 40-μm strainer, washed with PBS, and mechanically dissociated into single cells as previously described26. For enzymatic dissociation of brain tissue, previously published protocols were used12,13. Pelleted cells were suspended in FACS buffer (1% BSA in PBS) and stained on ice for 30 min with the following antibodies: rat anti-CD31-PE/CF594 (1:100, BD, 563616), rat anti-CD45-PE/Cy7 (1:200, Biolegend, 103114), mouse anti-heparan sulfate (1:100, Amsbio, clone 10E4, 370255-1), mouse anti-chondroitin sulfate (1:100, Sigma, clone CD-56, C8035), biotinylated HABP (1:150, Amsbio, AMS.HKD-BC41), fluorescein-conjugated SNA (1:300, Vector Labs, FL-1301-2), biotinylated MAAII (1:300, Vector Labs, B-1265-1), and StcE(E447D)–AF647 (5 μg ml−1). After washing cells with FACS buffer, secondary incubation with Alexa Fluor-conjugated streptavidin (1:1,000, Thermo Fisher Scientific) and secondary antibodies (1:400, Thermo Fisher Scientific) was carried out on ice for 20 min. Live cells were identified using Sytox Blue viability dye (1:1,000, Thermo Fisher Scientific, S34857). Flow cytometry analysis was performed on a BD LSRFortessa, and data were analysed using FlowJo software (TreeStar).
Microvessel imaging
Cerebral microvessels were isolated for immunofluorescence imaging using the same protocol described above for flow cytometry, except, instead of dissociating microvessels, they were fixed on 40-μm strainers with 4% PFA in PBS at room temperature for 15 min with gentle rocking. Microvessels were then washed with PBS and mounted on poly-d-lysine-coated slides (Thermo Fisher Scientific). Microvessels were blocked in 3% normal donkey serum (Jackson ImmunoResearch) with 0.3% Triton X-100 (Sigma) in TBS-T (1× TBS with 0.05% Tween-20) for 1 h, followed by 1 h incubation at room temperature with primary antibodies. For glyco-profiling of cerebral microvessels, the same primary antibodies, lectins, binding proteins and concentrations from our flow panel were used along with fluorescein-conjugated VVA (1:300, Vector Labs, FL-1231-2). Additional primary antibodies used include rabbit anti-C1GALT1 (1:100, Thermo Fisher Scientific, PA5-52814), rabbit anti-B3GNT3 (1:100, Thermo Fisher Scientific, PA5-21988), rabbit anti-CAV1 (1:100, Cell Signaling Technologies; 3267S), goat anti-CD31 (1:100, R&D, AF3628) and fluorescein-conjugated LEL (1:250, Vector Labs, FL-1171-1). For secondary staining, microvessels were washed three times with TBS-T for 5 min each, followed by incubation with Alexa Fluor-conjugated antibodies (1:250, Thermo Fisher Scientific) or streptavidin (1:1,000, Thermo Fisher Scientific) for 1 h at room temperature. Microvessels were then washed three times again and coverslipped with Vectashield Hardset Antifade Mounting Medium with DAPI (Vector Labs, H-1500-10) or ProLong Gold Antifade Mountant (Thermo Fisher Scientific, P36934). Imaging was performed on a confocal laser-scanning microscope (Zeiss LSM880). All observed single-plane microvessels were captured and then quantified using ImageJ software.
Immunofluorescence analysis
For luminal vascular labelling, mice were euthanized with 2.5% (v/v) Avertin and transcardially perfused via peristaltic pump at 2 ml min−1 with the following ice-cold solutions: 8 ml PBS, 10 ml of 5 μg ml−1 of StcE(E447D)–AF647 or SNA–Cy3 (Vector Labs, CL-1303-1), and 8 ml of 4% PFA. For all other immunofluorescence analysis, mice were euthanized with 2.5% (v/v) Avertin and manually perfused with PBS unless noted otherwise. Tissues were extracted and fixed in 4% PFA at 4 °C overnight before preservation in 30% sucrose in PBS. Tissues were sectioned into 40 μm slices using a microtome (Leica). Slices were subsequently blocked in 3% normal donkey serum with 0.3% Triton X-100 in TBS-T for 1.5 h at room temperature and incubated at 4 °C overnight with the following primary antibodies: goat anti-CD31 (1:100, R&D, AF3628), goat anti-Iba1 (1:100, Abcam, ab5076), rat anti-CD68 (1:100, Bio-Rad, MCA1957), goat anti-collagen type IV (1:100, Sigma, AB769), rat anti-NID1 (1:100, Thermo Fisher Scientific, MA1-06501), rabbit anti-CLDN5 (1:100, Thermo Fisher Scientific, 34–1600), mouse anti-ZO1 (1:100, Thermo Fisher Scientific, 33–9100), goat anti-albumin (1:100, Thermo Fisher Scientific, A90-134A), goat anti-PODXL (1:100, R&D Systems, AF1556). The following day, slices were washed three times with TBS-T, stained with the appropriate Alexa Fluor-conjugated secondary antibodies (1:250, Thermo Fisher Scientific) or Alexa Fluor-conjugated streptavidin (1:1,000, Thermo Fisher Scientific) for 2 h at room temperature, washed three times again, mounted, and coverslipped with Vectashield Hardset Antifade Mounting Medium with DAPI (Vector Labs, H-1500-10). Imaging was performed on a confocal laser-scanning microscope (Zeiss LSM880), and images were analysed using ImageJ. Luminal vascular coverage was calculated as vessel (CD31+ or COL4A+) area occupied by the marker of interest divided by total vessel area. Endothelial MFI calculated using CD31+ mask.
BBB leakage assay
Mice were anaesthetized and injected retro-orbitally with Sulfo-NHS-biotin (Thermo Fisher Scientific, 21335) at 0.25 mg g−1 body weight. The tracer was allowed to circulate for 5 min before perfusion with PBS. Hemibrains were post-fixed in 4% PFA overnight at 4 °C, cryopreserved in 30% sucrose, and sagittally sectioned into 40-μm slices. Sections were blocked and co-stained with CD31 and the appropriate secondary antibody as described earlier and Alexa Fluor 647-conjugated streptavidin (1:1,000, Thermo Fisher Scientific). Images were taken on a confocal laser-scanning microscope (Zeiss LSM880) and analysed using ImageJ software. Multicoloured gradient images were generated using the fire LUT in ImageJ. Permeability index of vessels was determined as the area occupied by tracer divided by the vessel area.
In vivo StcE treatment and cerebral bleeding assessment
All recombinant StcE proteins were produced as described22,23. For all applications, proteins were run through Pierce high-capacity endotoxin removal columns (Thermo Fisher Scientific) at least seven times following manufacturer’s instructions. Endotoxin levels were tested using HEK-Blue lipopolysaccharide (LPS) Detection Kit 2 (InvivoGen) according to manufacturer recommendations. Mice were injected retro-orbitally with 0.25 mg kg−1 StcE once a day for 2 days before perfusion with ice-cold PBS. Cerebral bleeding was visualized by eye post-perfusion and by H&E staining. For H&E staining, hemibrains and peripheral organs were formalin-fixed and paraffin embedded (FFPE) and cut into 5-μm-thick sagittal sections mounted on slides. Sections were deparaffinized in xylene (3 times, 3 min), hydrated in a series of graded alcohols (2× 100%, 1× 95%, 1× 80%; 3 min each), and stained with Richard Allan haematoxylin (4 min; followed by 30 s in 4% acetic acid in water and dipping in 0.3% ammonia water) and eosin with phloxine (30 s). Sections were then dehydrated (10 dips in 95% ethanol followed by 2× 1 min in 100% ethanol), cleared in xylene (3 times, 1 min), and coverslipped prior to imaging on a wide-field microscope (Zeiss AxioImager).
Luminex cytokine measurement
Plasma cytokine measurement was performed using the Luminex assay at the Human Immune Monitoring Center at Stanford University. The mouse 48-plex Procarta kit (Thermo, EPX480-20834-901) was used according to the manufacturer’s instructions. Plasma samples were diluted 1:3 and run in singlet on a 96-well plate alongside standard curve and quality control samples. Custom Assay Chex control beads (Radix BioSolutions) were added to all wells to assess nonspecific binding.
Luminal cerebrovascular proteome enrichment and peptide preparation
Young (3-month-old) and aged (21-month-old) mice (n = 6 mice per group) were euthanized with 2.5% (v/v) Avertin and transcardially perfused via peristaltic pump with the following ice-cold solutions: 8 ml of PBS, 20 ml of 0.5 mg ml−1 Sulfo-NHS-biotin (Thermo Fisher Scientific, 21217), and 10 ml of 50 mM Tris-PBS. Cerebral microvessels were isolated as described in earlier sections and collected for lysis via sonication in RIPA buffer (Thermo Fisher Scientific) supplemented with 1× protease inhibitor. Lysates were centrifuged at 13,000g for 15 min at 4 °C, and supernatant was saved for downstream enrichment. Protein concentration was measured by BCA (Pierce), and 1 mg of each sample was incubated with 70 μl of streptavidin magnetic beads (Thermo Fisher Scientific, 88817) at 4 °C overnight. The following day, beads were processed for proteomic analysis using a published protocol with some modificiations35. In brief, beads were washed once with 200 μl of 50 mM Tris-HCl (pH 7.5) and twice with 200 μl of 2 M urea in 50 mM Tris (pH 7.5) and incubated in 80 μl 2 M urea in 50 mM Tris with 1 mM dithiothreitol (Thermo Fisher Scientific, R0861) and 0.4 μg trypsin/LysC (Fisher, V5073) at 25 °C for 1 h shaking at 1,000 rpm. Supernatant was further reduced with 4 mM dithiothreitol for 30 min with shaking and alkylated with 10 mM iodoacetamide (IAA, Sigma, I1149) for 45 min in the dark with shaking. An additional 0.5 μg of trypsin/LysC was added to samples for overnight digestion at 37 °C with shaking. The following day, samples were acidified to 1% vol/vol formic acid and desalted using BioPureSPN mini C18 columns (Nest Group). All centrifugation steps were carried out at 50g for 1 min at room temperature. Columns were washed with 200 μl HPLC-grade methanol (Fisher) and equilibrated two times with 200 μl 0.1% formic acid in HPLC-grade water (Fisher) (solvent A). Samples were loaded onto columns, washed 4 times with solvent A, and eluted 2 times with 75 μl 0.1% formic acid in 80% acetonitrile (solvent B). The combined elutes were dried in a vacuum concentrator and reconstituted in 10 μl solvent A for LC–MS/MS.
Luminal cerebrovascular mass spectrometry-based proteomics
LC–MS/MS analysis was performed on a Q Exactive HF-X (Thermo Fisher Scientific) with an UltiMate 3000 RSLCnano system (Thermo Fisher Scientific). Peptides were loaded on an in-house 75-μm (inner diameter) capillary column packed with 40 cm of ReproSil-Pur 120 C18-AQ 1.9 μm resin (Dr. Maisch). Chromatographic separation was achieved using a flow rate of 300 nl min−1 with the following 120 min gradient: 96% A + 4% B for 18 min, 70% A + 30% B for 72 min, 60% A + 40% B for 15 min, and 4% A + 96% B for 15 min, where solvent A was 0.1 % formic acid in HPLC-grade water (Fisher) and solvent B was 0.1% formic acid in HPLC-grade acetonitrile (Fisher). Full MS scans were acquired at a resolution of 60,000, with an automatic gain control (AGC) target of 3 × 106, maximum injection time (IT) of 20 ms, and scan range 300–1,650 m/z in a data-dependent mode. MS2 scans were acquired with the following parameters: resolution of 15,000, AGC target of 1 × 105, maximum IT of 54 ms, loop count 15, TopN 15, isolation window 1.4 m/z, fixed first mass 100.0 m/z, normalized collision energy (NCE) 28 units, charge exclusion of unassigned, 1, 6–8 and >8, peptide match preferred, exclude isotopes on, and fragmented m/z values were dynamically excluded from further selection for a period of 45 s. Raw data were processed and analysed using MaxQuant and Perseus36. In brief, peptide spectral matches were made against a target-decoy Mus musculus reference proteome database downloaded from Uniprot. Methionine oxidation and N-terminal acetylation were specified as variable modifications, and carbamidomethylation of cysteines was specified as a fixed modification. Precursor ion search tolerance of 20 ppm and product ion mass tolerance of 20 ppm were used for searches. Both unique and razor peptides were used for quantification. Results were filtered to a 1% false discovery rate (FDR) at the peptide and protein levels. Proteins were quantified and normalized using MaxLFQ37 with a LFQ minimum ratio count set to 1. For quantitative comparative analysis, protein intensity values were log2-transformed, and missing values were imputed from a normal distribution with width 0.3 and downshift value of 1.8 using Perseus. Principal component analysis (PCA) was performed in Perseus using the Benjamini–Hochberg FDR with a cutoff of 0.05. GO term enrichments were performed using DAVID38 with the M. musculus proteome as a background.
Microvessel mass spectrometry-based proteomics
Brain microvessels were isolated and lysed in 1× RIPA buffer with 1× cOmplete protease inhibitor cocktail (Sigma) on ice. Lysates were centrifuged at 13,000g for 15 min at 4 °C, and supernatant protein concentration was measured by microBCA (Pierce). Samples were then reduced and alkylated by TCEP and CAA, followed by protein purification using SP3 (Cytiva). Samples were digested with 200 ng trypsin/LysC in pH 7.8 at 37 °C for 3 h, desalted, and eluted following the manufacturer’s instructions. Samples were acidified to 0.1% formic acid and filtered before loading onto the nanoLC system. One microgram protein was injected for all samples. LC–MS/MS analysis was performed on the TimsTOF Pro (Bruker Daltonics) coupled with the NanoElute system (Bruker Daltonics) with solvent A (0.1% formic acid in water) and solvent B (0.1% formic acid in acetonitrile). Tryptic peptides were loaded first on the trapping column Waters ACQUITY UPLC M-Class Symmetry C18 Trap Column, 100 A, 5 μm, 180 μm × 20 mm, and eluted with analytical column, IonOpticks Aurora Elite CSI 15 × 75 C18 UHPLC column. Elution gradient was set as 0 min 5% B; 9 min 12% B; 9.1 min 12% B; 27 min 30% B; 27.5 min 85% B and 36 min 85% B with a flow rate of 0.35 μl min−1 from 0 min to 9 min and reduced to 0.3 μl min−1 at 9.1 min until the end of the gradient. Eluted peptides were measured in diaPASEF mode with base method m/z range 100–1,700 and 1/k0 range of 0.85–1.30 V s cm−2. The source parameters were 1,400 V for capillary voltage, 3.0 l min−1 for dry gas, and 180 °C for dry temperature using Captive Spray (Bruker Daltonics). Collision energies (27 eV and 45 eV) were allocated for 1/K0 = 0.85 V s cm−2 and 1/K0 = 1.30 V s cm−2, respectively. Data were processed using Spectronaut (Biognosys AG, v19.1) for directDIA search with Swiss-Prot Mouse database downloaded on 3 March 2023. Default settings were used with a slight modification of minimum peptide length 6. Candidates were filtered using Q < 0.05 and absolute average log2 ratio ≥0.263.
RNA-seq glycosylation-related gene analysis
Previously published ageing and neurodegenerative disease RNA-seq datasets demonstrating robust brain endothelial cell enrichment were chosen for glycosylation-related gene analysis12,26,27. We filtered for glycosylation-related genes based on KEGG (Kyoto Encyclopedia of Genes and Genomes) listed glycosylation enzymes and related proteins. Most glycoproteins were excluded due to the enormous variety of members in this family which possess biological functions not directly relevant to glycosylation. Significantly upregulated and downregulated glycosylation-related genes in each dataset were used for Reactome pathway analysis with Padj < 0.05 set as the threshold for significant enrichment.
Bulk RNA-seq on bEnd.3 cells
The mouse brain endothelial cell line bEnd.3 (ATCC, CRL-2299) was cultured in high-glucose DMEM (Thermo Fisher Scientific, 10567022) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin and maintained in a humidified incubator containing 5% CO2 at 37 °C. For bulk RNA-seq analysis, bEnd.3 cells were grown in 6-well plates and treated with 5 nM StcE for 16 h at 37 °C. Cells were lysed and collected into RNAse-free Eppendorf tubes for total RNA extraction using the RNeasy Plus Micro kit (Qiagen, 74034). RNA quantity and quality were assessed by an Agilent 2100 Bioanalyzer (Agilent Technologies). All samples passed a high quality control threshold (RNA integrity number ≥9.7) and proceeded to cDNA library preparation by Novogene. Libraries were sequenced on the NovaSeq 6000 (paired-end, 2× 150 bp depth). Trimmed reads were aligned to the M. musculus reference genome GRCm38. Differential gene expression analysis and visualization were performed using DESeq2 (v1.32) (Supplementary Data 5). Genes with a Padj < 0.05 were used for GO biological pathway enrichment analysis.
bEnd.3 immunofluorescence analysis
For confocal imaging analysis, bEnd.3 cells were plated on round coverslips (EMS, 72196-12) in a 24-well plate and treated with 5 nM StcE for 16 h at 37 °C. Cells were fixed in 4% PFA for 15 min, blocked in 3% normal donkey serum with 0.3% Triton X-100 in PBS for 1 h at room temperature, and incubated at room temperature in blocking solution with the following primary antibodies for 1.5 h: goat anti-CD31 (1:100, R&D, AF3628), mouse anti-ZO1 (1:100, Thermo Fisher Scientific, 33–9100), rabbit anti-CAV1 (1:100, Cell Signaling Technologies; 3267S) and mouse anti-CLTC (1:100, Thermo Fisher Scientific, MA1-065). Cells were subsequently washed 3 times with PBS, stained with the appropriate Alexa Fluor-conjugated secondary antibodies (1:250, Thermo Fisher Scientific) for 1 h at room temperature, washed 3 times again, mounted, and coverslipped with Vectashield Hardset Antifade Mounting Medium with DAPI (Vector Labs, H-1500-10) or ProLong Gold Antifade Mountant (Thermo Fisher Scientific, P36934). Imaging was performed on a confocal laser-scanning microscope (Zeiss LSM880), and images were analysed using ImageJ.
For flow cytometry analysis, bEnd.3 cells were plated in a 24-well plate and treated with 5 nM StcE for 16 h at 37 °C. Cells were collected from plates using enzyme-free cell dissociation buffer (Thermo Fisher Scientific, 1315014) and resuspended in 1% BSA in PBS (FACS buffer). Cells were stained with the following antibodies on ice in FACS buffer for 30 min: APC-anti-CD54 (1:100, BioLegend, 116120), FITC-anti-VCAM1 (1:100, Thermo Fisher Scientific, 11-1061-82), Alexa Fluor 647-anti-CD62P (1:100, BD Biosciences, 563674), anti-CD62E (1:100, Thermo Fisher Scientific, 14-0627-82) followed by Alexa Fluor 555-conjugated anti-mouse IgG (1:400, Thermo Fisher Scientific). Live cells were identified using Sytox Blue viability dye (1:1,000, Thermo Fisher Scientific, S34857). Flow cytometry analysis was performed on a Sony SH800S sorter, and data were analysed using FlowJo software (TreeStar). Only live, singlet cells were used for analysis of adhesion molecule MFI.
ROS assays
For in vitro ROS assays, bEnd.3 cells were plated in 8-well µ-slides (Ibidi, 80826) and treated with 5 nM StcE, 5 μg ml−1 of LPS, or an equivalent volume of saline in complete medium for 16 h at 37 °C. Cells were then washed two times with PBS, and cellular ROS assays were carried out according to manufacturer instructions. In brief, 20 µM of DCFDA (Abcam, ab113851) and 1× ROS Deep Red Dye (Abcam, ab186029) were used for their respective assays and incubated with cells for 30 min at 37 °C in the dark. Cells were washed three times with PBS and imaged immediately on a confocal laser-scanning microscope (Zeiss LSM880). Quantification of MFI was performed using ImageJ software. For in vivo ROS assays, cerebral microvessel were isolated as described earlier and incubated with 1× ROS Deep Red Dye (Abcam, ab186029) for 30 min at 37 °C in the dark. Microvessels were then washed with 5 ml of PBS and pelleted by centrifugation at 2,000g for 10 min and transferred to 8-well µ-slides (Ibidi, 80826) for immediate imaging on a confocal laser-scanning microscope (Zeiss LSM880). All observed single-plane microvessels were captured and then quantified using ImageJ software.
Western blot
Whole-brain microvessel lysates (30 µg) were boiled in 1× NuPAGE LDS Sample Buffer (Thermo Fisher Scientific) and 5% 2-mercaptoethanol (Sigma) at 95 °C for 10 min. Lysates and the Precision Plus Protein Dual Color Standards (Bio-Rad, 1610374) were loaded onto a 4–12% NuPAGE Bis-Tris precast gel (Thermo Fisher Scientific) and run in NuPAGE MOPS buffer (Thermo Fisher Scientific) at 160 V for 1 h. The gel was transferred to a 0.45-µm nitrocellulose membrane (Bio-Rad) using the Mini Trans-Blot Cell (Bio-Rad) and NuPAGE transfer buffer (Thermo Fisher Scientific) at 100 V for 1.25 h. Protein transfer was verified using Ponceau S staining for 5 min followed by de-staining in de-ionized water. The membrane was blocked with 5% non-fat dry milk (Bio-Rad) in TBS-T for 1 h at room temperature and then incubated with IRDye 800CW streptavidin (1:2,500, LI-COR Biosciences) in 5% non-fat dry milk (Bio-Rad) in TBS-T for 1 h at room temperature. The membrane was then washed three times with TBS-T for 5 min each and imaged on the LI-COR Odyssey DLx.
Design and cloning of pAAV-sCLDN5 constructs
The Ple261 MiniPromoter (‘sCLDN5’) pEMS1938 was a gift from E. Simpson (Addgene plasmid #82563). A cis rAAV genome plasmid with AAV2 inverted terminal repeats was utilized for cloning of a sCLDN5 and EGFP reporter using restriction enzymes and In-Fusion Snap Assembly (Takara Bio). To knock down C1galt1 in brain endothelial cells, de novo predictions of small interfering RNA (siRNA) guides targeting C1galt1 were generated using the DSIR algorithm39 and subsequently filtered using ‘Sensor rules’ to select for sequences with highly favourable small hairpin RNA (shRNA) features40,41. Three de novo 97-mer miR-E shRNA sequences (Supplementary Table 2) were synthesized (IDT) and inserted into pAAV-sCLDN5-EGFP using restriction enzyme cloning for in vitro evaluation. To overexpress C1GALT1 and B3GNT3 in brain endothelial cells, P2A-C1GALT1 and P2A-B3GNT3 were cloned into pAAV-sCLDN5-EGFP using restriction enzyme cloning to generate pAAV-sCLDN5-EGFP-P2A-C1GALT1 and pAAV-sCLDN5-EGFP-P2A-B3GNT3, respectively.
In vitro pAAV evaluation
bEnd.3 cells were split into a 24-well plate 1 day before transfection. Cells at approximately 70% confluency were transfected with the pAAVs described in the previous section via Lipofectamine 3000 according to the manufacturer’s recommendations (Thermo Fisher Scientific, L3000008). After 16 h, the cell medium was replaced with fresh, pre-warmed medium. Then, 2.5 days after transfection, cells were collected from plates using enzyme-free cell dissociation buffer (Thermo Fisher Scientific, 1315014) and resuspended in FACS buffer. Cells were stained with StcE(E447D)–AF647 (5 μg ml−1) on ice for 20 min. Live cells were identified using Sytox Blue viability dye (1:1,000, Thermo Fisher Scientific, S34857). Flow cytometry analysis was performed on a Sony SH800S sorter, and data were analysed using FlowJo software (TreeStar). Only live, singlet, EGFP+ cells were used for analysis of StcE(E447D)–AF647 MFI.
AAV production
For production of PHP.V1-sCLDN5::EGFP and all other plasmids containing the PHP.V1 (pUCmini-iCAP-PHP.V1 was a gift from V. Gradinaru; Addgene plasmid #127847) capsid, AAV production was performed in-house utilizing a previously published protocol42. In brief, triple transfection of HEK293T cells was performed on 90–95% confluent cells in DMEM containing Glutamax supplemented with 5% FBS and non-essential amino acids. Fresh, warm medium was replaced 12 h post-transfection. Medium was collected 72 h post-transfection. Fresh, warm medium was added and collected along with cells 120 h post-transfection and combined with the previous fraction. Cells and medium were centrifuged at 2,000g for 15 min at room temperature. Supernatant was collected in a separate bottle and combined with 40% (wt/vol) PEG (final concentration 8% wt/vol PEG) and incubated on ice for 2 h before transferring to 4 °C overnight. Cell pellet was resuspended in buffer containing salt-active nuclease (SAN, ArcticZymes Technologies) and incubated at 37 °C for 1 h before transferring to 4 °C overnight. PEG medium was centrifuged at 4,000g for 30 min at 4 °C. After centrifugation, supernatant was bleached and discarded. PEG pellet was resuspended in SAN + SAN buffer, combined with the previous fraction, and incubated at 37 °C for an additional 30 min. Lysate was centrifuged at 2,000g for 15 min at room temperature, and supernatant was loaded onto an iodixanol gradient (15%, 25%, 40% and 60% fractions). Gradients were transferred to an ultracentrifuge (Beckman Coulter) using a Type 70 Ti rotor set at 350,000g for 2 h and 25 min at 18 °C. AAV particles were collected from the 40/60% interface, washed in PBS, and concentrated using an Amicon Ultra-15 (Millipore Sigma) filter device with a 100 kDa cutoff. AAV titration was performed using the AAVpro Titration Kit (for Real Time PCR) Ver.2 (Takara Bio). AAVs were injected retro-orbitally at 8 × 1011 viral genomes per mouse.
snRNA-seq of mouse brain tissue
Pooled cortical and hippocampal tissue were dissected from frozen brain hemispheres from young (5-month-old) and aged (19-month-old) mice that had been injected with AAVs 8 weeks prior to takedown. Brain tissue was minced using a razor blade on ice and transferred to 2 ml glass Dounce tissue grinders (Sigma, D8938) with 2 ml EZ lysis buffer (Sigma, NUC-101) on ice. Tissues were homogenized using 25 strokes with pestle A followed by 25 strokes with pestle B, incorporating a 180° twist. Tissue homogenate was incubated on ice for 5 min and then centrifuged at 500g for 5 min at 4 °C. The pellet was resuspended with 4 ml EZ lysis buffer, incubated on ice for another 5 min, and pelleted as before. The pellet was then resuspended with chilled PBS and filtered through a 35-µm strainer before blocking with Mouse Fc block (1:50, BD 553142) in FACS buffer for 5 min. Nuclei were subsequently stained with AF647-anti-NeuN (1:100, Abcam, ab190565) in FACS buffer with 0.2 U µl−1 RNase inhibitor (Takara, 2313 A) for 30 min on ice, washed with FACS buffer, and resuspended in FACS buffer with Hoechst dye (1:2,000, Thermo Fisher, H3570) and 0.2 U µl−1 RNase inhibitor (Takara, 2313 A). Using a Sony MA900 Cell Sorter, approximately 50,000 singlet nuclei were sorted into 1.5 ml Eppendorf tubes containing 10 mg ml−1 UltraPure BSA (Thermo Fisher, AM2618) and 0.2 U µl−1 RNase inhibitor (Takara, 2313 A) in PBS. Collected nuclei were centrifuged at 400g for 5 min at 4 °C with slow deceleration. Supernatant was removed leaving about 40 µl of suspended nuclei. Nuclei were counted using a haemocytometer (Sigma, Z359629) and assessed for concentration and quality. snRNA-seq libraries were prepared using the Chromium Single Cell 3′ Kit v3.1 (10X Genomics) according to the manufacturer’s protocols, targeting 10,000 nuclei per sample. Twelve polymerase chain reaction (PCR) cycles were applied to generate cDNA, and eleven PCR cycles were applied for final library generation. Quality control of cDNA and libraries were conducted using a High Sensitivity D5000 ScreenTape (Agilent). The final snRNA-seq libraries were sequenced on a NovaSeq X.
snRNA-seq quality control
Gene counts were obtained by aligning reads to the M. musculus reference genome GRCm38 using CellRanger software (v4.0.0) (10X Genomics). Ambient RNA was removed from each sample using SoupX (v1.6.2) and droplets containing multiple nuclei were filtered out using DoubletFinder (v2.0.4). We used Seurat (v4.1.1) to further exclude cells with fewer than 200 or more than 5,000 features and cells with more than 10% mitochondrial genes. In total, 69,250 nuclei remained and were used for further analysis (Supplementary Fig. 3a–d).
Normalization and cell clustering
Each sample was normalized using Seurat’s SCTransform function, and samples were integrated using Seurat’s anchoring and integration functions. PCA was then run on the integrated dataset and the first fifteen principal components were used to generate a shared-nearest-neighbours graph. Clustering of nuclei was determined using the Louvain algorithm with a resolution of 0.3. UMAP was performed using the first 15 principal components and 30 nearest neighbours. Clusters were annotated for cell type using common cell-type markers identified by the FindConservedMarkers function applied to each cluster (Supplementary Fig. 4a; Supplementary Data 4).
Pseudobulk differential gene expression
Pseudobulk counts were derived by aggregating raw counts for each sample using Seurat’s AggregateExpression function. DESeq2 was then used to perform bulk data normalization and differential gene expression across groups using default parameters. P value correction was carried out using the Benjamini–Hochberg procedure (FDR = 0.05) for each comparison. Genes with FDR <0.05 were used for subsequent KEGG pathway enrichment and Metascape43 analysis (Supplementary Data 5).
Behavioural assays
In the Y maze test, the Y maze is made up of 3 white, opaque plastic arms at 120° angles from each other. At the beginning of the trials, mice were placed in the end of 1 arm and allowed to freely explore all 3 arms for 5 min. An arm entry was defined as having all four limbs inside an arm. The maze was cleaned with 70% ethanol between animals and before the first animal to eliminate traces of odour. The number of arm entries and triads (a set of consecutive arm entries) were recorded. The spontaneous alternation was calculated by dividing the number of triads by the number of possible alternations multiplied by 100.
In fear conditioning tests, mice were trained to associate cage context and an audiovisual cue with an aversive stimulus (foot shock). On day 1 (training), mice were placed in a cage and exposed to two periods of 30 s of paired cue light and 1,000-Hz tone followed by a 2-s foot shock (0.6 mA), with a 180-s interval. On day 2 (contextual fear conditioning), mice were re-exposed to the same cage context and freezing behaviour was recorded during minutes 1.5 to 6 using a FreezeScan tracking system (Cleversys). On day 4 (cued fear conditioning), mice were placed in a novel context that contained different odour, floor texture, and chamber walls and re-exposed the same cue light and tone from day 1 after 2 min of exploration. Freezing behaviour was recorded for 1.5–6 min following the cue using the FreezeScan tracking system (Cleversys).
Statistical analysis
Data handling and statistical analyses were performed using R studio (v4.1.1) and GraphPad Prism Software unless stated otherwise. All statistical analyses comparing measurements between two groups were carried out using unpaired two-tailed Student’s t-tests. All statistical analyses comparing measurements between three or more groups were carried out using one-way ANOVA tests with post hoc tests for multiple comparisons. A P value of <0.05 was considered significant. All experimental procedures involving multiple experimental groups were carried out in an alternating fashion when possible to, avoid temporal and technical biases. Data in Figs. 1b–d,h–o, 2a,b,f–j, 3e–n, 4f–i and 5j–l and Extended Data Figs. 2a, 3f–i, 7a–c,h,i and 8h–k were successfully replicated in at least two independent experiments.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.