Compilation of genetic diversity change measures
We conducted a literature search of peer-reviewed publications using the Web of Science (WOS) advanced search functions to find published works that contained temporal measures of genetic diversity. Our search was intentionally broad and followed established preferred reporting items for systematic reviews and meta-analysis (PRISMA) protocols as closely as possible37,38. The online search was conducted on 18 January 2019 using keywords targeting population genetic measurements, regardless of the direction of change (for example, ‘increase’ and ‘decrease’ were both included as search terms) (Supplementary Information 2.1 and Supplementary Data 4). A total of 80,271 records were retrieved, 78,727 after duplicate removal. We obtained full texts for 70,069 of these records. The remaining records were screened manually via their titles, abstracts and keywords, of which 8,596 were excluded against our inclusion criteria; 62 full texts could not be obtained (Extended Data Fig. 2 and Supplementary Information 2.1). We then performed full text mining in R v.3.5.239, using the packages pdfsearch v.0.2.340, dplyr v.0.8.041, and stringi v.1.3.142 to remove records that did not contain population genetic keywords (Supplementary Information 2.2). This resulted in 34,346 putatively includable studies of genetic diversity change, which were classified into thematic clusters using the package revtools v.0.4.043 (Supplementary Information 2.2).
We performed initial screening and data extraction for all 34,346 works, followed by a series of re-extraction and data validation steps. Manual screening of studies against inclusion criteria, and extraction of genetic diversity measurements and metadata took place simultaneously by members of the authorship team, via multiple workshops using shared written guidelines (Extended Data Tables 1 and 2 and Supplementary Information 2.3–2.6). Studies were suitable for inclusion in our analysis only if they satisfied all of the following criteria:
-
The research must report primary, quantitative, empirical data from a multicellular nonhuman organism.
-
Laboratory and experimentally manipulated populations were excluded, where these experimental manipulations were for the purpose of testing a hypothesis related to population demography or genetics (note that populations established in controlled conditions for supportive breeding or propagation were potentially includable, such as ‘captive’ or ‘agricultural’ populations),
-
The time frame of the study plausibly took place over timescales likely to have been impacted by human activities, regardless of whether the study organism was actually impacted by human activities (in general, we targeted genetic diversity changes in the last few hundred years and excluded studies on ancient admixture or expansion in response to events on ‘geological’ timescales; further detail in Supplementary Information 2.4),
-
The study design enabled a temporal comparison of population genetic measurements (for example, samples collected over multiple years) or inference (for example, coalescent genetic studies),
-
The study reports a quantitative measurement of within-population ‘genetic diversity’ (broadly defined), and an associated measurement error and sample size. Genetic diversity statistics were obtained from main texts (including tables and figures) and supplementary materials, but no re-analysis of published datasets was conducted. Summary statistics (mean and s.d.) were calculated from tabulated data where available.
In addition to recording bibliographic data for each record, we extracted data that would enable us to calculate our effect sizes (see below and Supplementary Information 2.4), as well as corresponding metadata for meta-regression (Extended Data Tables 1 and 2 and Supplementary Information 2.5 and 2.6). Our dataset included many studies for which the main goal was not an assessment of genetic diversity or change in genetic diversity per se, but which nevertheless reported temporal measures of genetic diversity that otherwise met the inclusion criteria for our meta-analysis.
We captured genetic diversity statistics aligned with three possible study designs (see Supplementary Information 2.4): (1) linear measure of change (for example, regression of two or more time points), yielding primarily statistical measurements, such as regression coefficients or t statistics; (2) two timepoint comparison (for example, comparison of two means), yielding primarily pairs of mean diversity estimates; or (3) coalescent analysis, yielding either statistical or genetic measurements, obtained by probabilistic modelling of past effective population sizes using a single sample in time44,45. The latter were uniquely identified in our analysis due to important differences in the underpinning theoretical framework for coalescent analyses. That is, ‘early’ measures of genetic diversity are not taken from real biological samples, but instead inferred from recent data and principles of genetic inheritance. Further, early time points in coalescent analyses may be identified by authors a priori based on environmental or other non-genetic hypotheses, or post hoc on the basis of substantive patterns in the data.
Genetic diversity change data were recorded alongside corresponding error estimates and sample sizes; all were extracted using the same level of precision as reported in the paper. We also recorded the time frame of the study (early and recent years, used to calculate study duration and study midpoint), plus amount and type of genetic data used (for example, number of loci, genetic marker type, genome). We classified genetic diversity change data into four metric types, aligned with ‘essential biodiversity variables’ for genetic composition28: (1) variant counts (for example, allelic richness); (2) evenness of variant frequencies (for example, expected heterozygosity and nucleotide diversity); (3) population means of individual-level variation (for example, observed heterozygosity and pedigree inbreeding); and (4) integrated statistics (for example, effective population size; see below).
We were particularly interested in associations between genetic diversity change and ecological disturbance or conservation management action, so these ‘impact metadata’ were collected where threats and/or conservation management actions were reported in a paper as plausibly impacting the study population between the sampling time points of the study. In principle, we categorized ecological disturbances as events with potential to impair conditions for the focal species or its habitat, and conservation management actions as human activities intended to improve conditions for the focal species or its habitat. For the latter, we also considered the intensity of conservation management actions (that is, the magnitude of conservation intervention as probably experienced by the focal species). We also collected additional metadata, as the objectives of ecological disturbances are likely to vary across species. For example, disturbance is often intentional for pests and pathogens (for example, population reduction), whereas disturbance of threatened species can include indirect or unintentional consequences of human activity (for example, habitat loss and fragmentation). Brief definitions of the categories that we used for each of these variables are in Extended Data Table 1 and 2 and full definitions are provided in Supplementary Information 2.5.
Additional moderators that were collected included (Supplementary Information 2.6): taxonomic identity of study species (nomenclature standardized by literature review to align with Open Tree of Life46), country and terrestrial and/or marine realm of the locality where samples were collected, following refs. 36,47,48, along with unique site identifiers in the case of multiple populations reported in a publication. We also collected the threat status of the study species30; generation length of the study species (Supplementary Data 5); and classification of domesticated species or populations considered as pathogens or pests (based on description in the source publication, relevant databases or other published sources).
Many studies reported multiple measurements of genetic diversity that were suitable for inclusion in our analysis. We extracted independent measures of genetic diversity change per publication taking into consideration the sampling scheme of the reported study and analysis of data subsets (Supplementary Information 2.4). Procedures for controlling non-independent data, missing data, infinite confidence intervals (in estimates of effective population size), zero variances, and unconventional study designs are described at Supplementary Information 2.7–2.9. After initial extraction, all included studies were re-processed by at least two additional members of a small validation group from the authorship team, to ensure consistency in the collection of genetic and metadata (Supplementary Information 2.10).
Systematic review
Full bibliographic details for each included study were automatically downloaded from the WOS during the original search (see also Supplementary Data 1–3). Additional bibliographic metadata were also collected from the WOS, including journal title abbreviations, WOS subject categories for which each journal ranked highest, and the impact factor percentile ranking for each journal within its WOS category for 2020. Publication trends and the characteristics of studies included in our final dataset were summarized visually using the R packages ggplot2 v.3.4.349, treemapify v.2.5.550 and ggridges v.0.5.451. We also explored patterns of co-occurrence and characterized the variation of ecological disturbance and conservation management actions reported across our full dataset, and data subsets of the five most data-rich taxonomic classes. We calculated pairwise Spearman’s rank correlation coefficients (r) among ecological disturbance categories, and conservation management actions, and visualized results in R using corrplot v.0.9252. We also examined relationships among these variables using principal component analysis, visualized with the package factoextra v.1.0.753.
Phylogeny
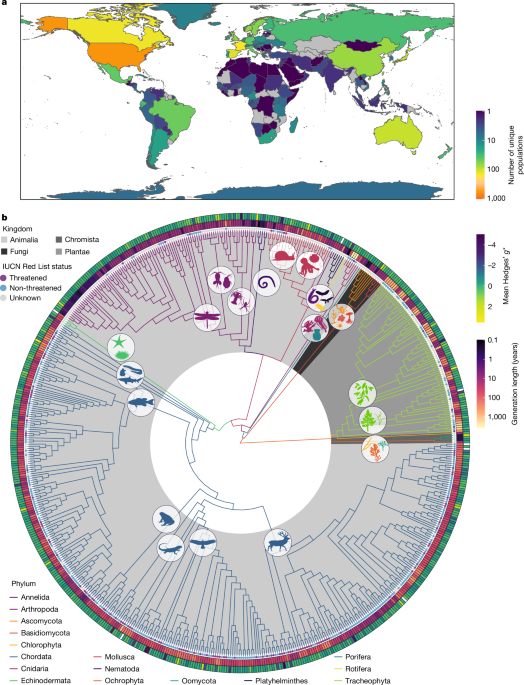
To visualize the taxonomic diversity of species in our dataset and their evolutionary relationships, we generated a phylogenetic tree using the R package rotl v.3.0.1254 using Open Tree of Life IDs as described above. Saccharomyces cf. cerevisiae (ott id 7511391) was used as the outgroup. Six species could not be placed in the phylogeny due to unresolved taxonomy: the Japanese mud snail (Batillaria attramentaria), white seabream (Diplodus sargus), a fruit fly (Drosophila pseudoobscura), a sea snail (Euparthenia bulinea), fourfinger threadfin (Eleutheronema tetradactylum) and the bicolour damselfish (Stegastes partitus). The phylogeny was visualized using the R packages ggtree v.3.8.255, ggtreeExtra v.1.10.056, ggimage v.0.3.357 and rphylopic v.1.2.158. Silhouettes of representative organisms for each taxonomic class were downloaded from PhyloPic (https://www.phylopic.org; see Supplementary Information 2.6 for credits). Owing to the taxonomic diversity of species in our study, obtaining a dated tree across all species was not possible and so the topology of the tree was used in modelling.
Effect size extraction and calculation
For each comparison that satisfied our inclusion criteria, we calculated Hedges’ g* (sometimes referred to as Hedges’ d59 with sample size correction J) as our measure of effect size. Hedges’ g* was selected as the effect size measure as it is based on the standardized mean difference between two values, in our case the ‘early’ and ‘recent’ time points, minimizes over-inflation of effect size estimation in studies with sample sizes <20, and outperforms other common effect size measures such as Cohen’s d and Glass’ Δ when the assumption of homogeneity of variance is violated60. All formulae used to evaluate Hedges’ g* and its error are reported in Supplementary Information 2.11.
Calculation of Hedges’ g* requires the sample size and error associated with the measure of genetic diversity change. Depending on the way in which genetic diversity metrics or their summary statistics are calculated, the associated sample size for effect size calculation may be for example, the number of loci, the number of samples, the number of populations or a rarefied sample size. For comparisons based on linear measures of genetic diversity change, which varied in the methods used to determine genetic change, each paper was manually checked to retrieve the appropriate sample size and error. For two timepoint comparisons and comparisons based on coalescent analyses, we followed a hierarchical procedure to establish the sample size to use for each effect size (Supplementary Information 2.11). Multiple error types were reported (for example, s.d. or confidence intervals), and so Hedges’ g* was calculated using published formulae for interconversion of these data types (Supplementary Information 2.11).
For comparisons where an effect size was calculated, the direction of the effect was determined. We ensured consistent directionality among the following measures of genetic diversity (note that many metrics were recorded in our dataset, and so the abbreviations reported below are summaries only):
-
(1)
Variant counts were all positively associated with genetic diversity: mean of alleles across loci (A), standardized by sample size (AR), sum of alleles across loci (TA), total number of private alleles (pA) and number of polymorphic loci (NPL).
-
(2)
Variant frequencies:
-
a.
Positive: expected heterozygosity (HE), nucleotide diversity (π), haplotype diversity (h), Shannon diversity index (H), polymorphic information content (PIC), number of effective alleles (NEA), frequency of an allele of interest (Freq) and mean individual nucleotide p-distance (NPD, occasionally seen in major histocompatibility complex and similar studies).
-
b.
Negative: population-level inbreeding coefficient or selfing/outcrossing rate (FIS), mean relatedness or kinship among individuals (R), band sharing score (BS) and among-population FST (apFST).
-
a.
-
(3)
Individual-level diversity measures:
-
a.
Positive: observed heterozygosity (HO), standardized observed heterozygosity (SH) and mean number of alleles per individual (Ai).
-
b.
Negative: individual-level inbreeding coefficient or coancestry (F).
-
a.
-
(4)
Integrated statistics were all positively correlated with genetic diversity: effective population size (Ne), effective number of breeders (Nb), female effective population size (Nf), effective population size estimated from demographic data (Nd), effective population size estimated from a population census, and calculated based on an assumption about the ratio between effective and census population sizes (Nc).
For comparisons where genetic diversity metric type was recorded as ‘other’, each paper was manually checked to determine the correct direction of the effect given the context of the metric within the publication and the authors’ interpretation of genetic diversity change as a loss or gain. For negatively correlated metrics, we multiplied the Hedges’ g* effect size by −1 to reverse the direction of the effect—that is, across our dataset a positive Hedges’ g* represents an increase in genetic diversity and a negative Hedges’ g* represents a loss of genetic diversity.
All calculated effect sizes >|4| were manually examined as potential outliers by a single member of the research team, to confirm absence of data entry errors. Considering our wide diversity of statistics, these data were also checked for possible calculation errors, misinterpretation of statistical error (for example, standard error versus s.d.), or other discrepancies. Results of screening of extreme values can be found in Supplementary Information 1.4.
Meta-analysis
We fitted multi-level Bayesian hierarchical models in the R package MCMCglmm v.2.3461, with paper ID as a random effect for all models to account for non-independence introduced by studies that report multiple, includable effect sizes. Genetic diversity change was modelled per generation by including the z-standardized number of generations for that species (that is, number of years passed between the early and recent time points, divided by generation length) as a fixed effect in all meta-regressions. Unless otherwise stated in Supplementary Information 2.12, all meta-regressions also included a fixed effect of the z-standardized study midpoint (year).
Each model was run for 6,000,000 iterations, with a burn-in of 200,000 and a thinning interval of 5,000, using the weakly informative inverse-gamma prior. We report the posterior mean and the 95% HPD credible intervals for each model set. Estimates with a 95% HPD credible interval excluding zero were considered statistically significant at α = 0.05. Model diagnostics were visually checked for no pattern in the trace plots; effective size >1,000 and autocorrelation <0.1 between lag points were both checked using coda v.0.19.462. Chain convergence was confirmed by passing the Heidelberger and Welch’s half-width and stationarity tests in coda. Additionally, each model was independently run three times to calculate a Gelman-Rubin convergence diagnostic of <1.1 using the potential scale reduction factor. The deviance information criterion (DIC) was obtained for each of the three models, and the model with the lowest DIC was selected for interpretation.
Our base model included fixed and random effects described above (that is, fixed effects = z-standardized midpoint, z-standardized number of generations; random effect = paper ID), although variations of this model underwent sensitivity testing to determine the influence of including phylogeny as an additional random effect, and including extreme values in the dataset (described in Supplementary Information 2.11). The extended heterogeneity statistic63 was calculated for both the base model and the sensitivity testing model that included phylogeny. Extended heterogeneity statistics partition total heterogeneity (I2total) into phylogenetic variance (in the phylogenetic model, I2phylogeny), study ID variance (I2study) and residual variance63,64 (I2residual). For the phylogenetic model, we also obtained lambda (phylogenetic signal (H2)) as the variance of the random effect of phylogeny divided by the total variance of all random effects (phylogeny, study ID and residual variance). Total heterogeneity was high, but phylogenetic signal only explained 5.48% of overall variance, so was excluded from further modelling (see also Supplementary Information 1.5); this also allowed for simplification of the model structure. Based on the results of the sensitivity testing (Supplementary Information 1.5), we excluded phylogeny and extreme values from subsequent meta-analytic modelling.
We assessed publication bias in our meta-analysis using two methods. First, we investigated time-lag bias, where different patterns in genetic diversity change may be reported over the years of publication. Such bias may plausibly occur given that methods for measuring genetic diversity have advanced substantially in recent decades. Therefore, we fitted the final base model with the addition of a z-standardized year of publication fixed effect. Evidence of time-lag bias is inferred if the 95% HPD credible interval of the slope estimate excludes zero. Second, we plotted Hedges’ g* precision against Hedges’ g* in a funnel plot to visualize possible publication bias that can occur if, for example, smaller studies without statistically significant results are not published. We did not observe time-lag bias nor funnel plot asymmetry (Supplementary Information 1.5). Given the high heterogeneity and lack of detectable publication bias in our dataset, we proceeded with meta-regression modelling.
Meta-regressions were conducted to assess the impact of different moderator variables on genetic diversity change. These were broadly categorized into moderators related to: (1) how genetic diversity change is measured (that is, study design); (2) where genetic diversity change is measured and in what species (that is, population context); (3) ecological disturbances (that is, threats); and (4) conservation interventions (that is, conservation management). In meta-regression, the coefficients estimate how each category differs from the nominated reference group, represented by the intercept65. As all moderator variables were categorical, we performed separate meta-regressions for each moderator to avoid the confounding effects of correlations and allow for biologically meaningful interpretation of categorical variables relative to the intercept. For all models, moderator variables were only included if there were 10 or more effect sizes contributing to a category65. All models were run with the weakly informative inverse-gamma prior, the paper ID random effect and the standardized year midpoint and the number of generations over which the study took place (as a measure of study length) as fixed effects (unless otherwise specified), and additional fixed effects described in Supplementary Information 2.12.
Inclusion and ethics statement
No ethical approval or guidance was required as data were collected only from previous studies.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.