Trypanosome culture and genetic manipulation
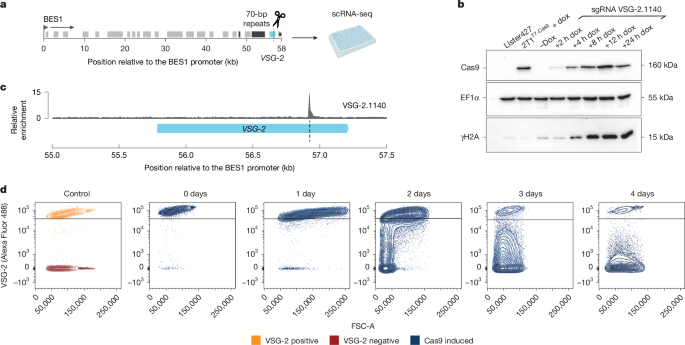
Bloodstream form T. brucei Lister 427, P10 and N50 (ref. 35), 2T1T7 (ref. 40) and 2T1T7 Cas9 trypanosomes along with their derivatives, were maintained at 37 °C and 5% CO2 in HMI-11 supplemented with 10% fetal calf serum and the appropriate selective drug41. Cells were maintained below 2.0 × 106 cells per ml. Cells were electroporated as previously described42. The 2T1T7 Cas9 cell line was generated by transfecting 2T1T7 cells with the pRPaCas9 plasmid43, which integrates into a tagged ribosomal DNA (rDNA) spacer region40. Cas9 expression was induced with 1 µg ml−1 doxycycline. For the SL-Smart-seq3xpress switching assays, Cas9 induction was maintained throughout the time course. To generate clonal populations of VSG switched cells following the induction of a DSB, the induced population was diluted to 8–10 cells per ml in HMI-11 and spread across the wells of a 96-well plate in 100-µl volumes. Plates were left for 5 days for the cells to recover. Cells were counted with a Beckmann Coulter cell counter.
Sanger sequencing of expressed VSG transcripts
RNA was extracted from 5.0 × 106 cells using the NucleoSpin RNA Kit (Macherey & Nagel) according to the manufacturer’s instructions. RNA was stored at −80 °C. First strand cDNA was synthesized from 5 µg of the extracted RNA using the SuperScript II Reverse Transcriptase (Invitrogen) and the oligo(dT)12-18 primer as per the manufacturer’s instructions. First strand cDNA was stored at −20 °C. Expressed VSG transcripts were amplified from the first strand cDNA by PCR using a forward primer specific to the SL sequence (5′-GACTAGTTTCTGTACTAT-3′) and a reverse primer specific to the conserved 3′ sequence of VSG mRNAs (5′-CCGGGTACCGTGTTAAAATATATC-3′). For each PCR reaction, 1 µl of 1:50 diluted first strand cDNA was used. PCR products were visualized following agarose gel electrophoresis and the VSG amplicon purified from the agarose using the Nucleospin Gel and PCR Clean-up Kit (Macherey & Nagel) as per the manufacturer’s instructions. Sanger sequencing of at least 100 ng of purified PCR product was performed by Eurofins Genomics using either of the primers used in the VSG amplification PCR.
sgRNA design and cloning
sgRNA target sequences were designed with Protospacer Workbench44 (v.0.1.0 beta) using the Lister 427 2018 genome assembly (ref. 10, https://tritrypdb.org/) as the reference database and optimized for use with SpCas9. sgRNA target sequences were selected according to their Bowtie Score (a measure of off-target Cas9 activity) and the Doench-Root-Activity score (a measure of sgRNA activity). As described in ref. 43, ‘aggg’ was added to the forward primer sequence and ‘caaa’ to the reverse primer sequence to create the BbsI cloning sites. The target sequences were cloned into the pT7sgRNA plasmid and transfected into the 2T1T7 Cas9 cell line as previously described in ref. 43. The pT7sgRNA plasmid integrates into a random rDNA spacer region.
Western blot analysis of Cas9 and γH2A expression
Total protein extract from 2.0 × 106 cells was boiled in 1× lysis buffer (1:3 4× Laemmli:1× RIPA, 2 mM dithiothreitol (DTT), 1% β-mercaptoethanol) and separated on a 12.5% SDS–PAGE gel. Separated proteins were transferred onto a methanol-equilibrated PVDF (polyvinyl difluoride) membrane using a Bio-Rad Mini Trans Blot Cell according to the manufacturers’ instructions. To visualize transferred proteins, the membrane was stained with 0.5% Amido black solution (in 10% acetic acid). Destaining was performed with 1× destaining solution (25% isopropanol, 10% acetic acid). The blotted PVDF membrane was cut into three according to the prestained protein ladder before blocking: above 70 kDa for the detection of SpCas9, between 70 and 25 kDa for the detection of the EF1α loading control and below 25 kDa for the detection of γH2A. Cas9 and EF1-α blots were blocked in 5% milk/PBS-T and γH2A blots were blocked in 3% BSA/PBS-T at room temperature. The membranes were washed three times with 1× PBS-T and primary antibody incubation was performed overnight at 4 °C. The primary antibodies were used at the following dilutions: anti-Cas9 (1:1,000 in 5% milk/PBS-T, Active Motif, clone 7A9-3A3); anti- EF1α (1:20,000 in 1% milk/PBS-T, EMD Millipore Corporation, clone CBP-KK1) and anti-γH2A (1:200 in 1% milk/PBS-T, from L. Glover, Institut Pasteur). After washing the membranes three more times with PBS-T, the following horseradish peroxidase-conjugated secondary antibodies were used: for Cas9, anti-mouse (1:10,000 in 1% milk PBS-T, GE Healthcare, code NA931V); for γH2A, anti-rabbit (1:2,000 in 1% milk/PBS-T, GE Healthcare, code NA934V) and for EF1-α, anti-mouse (1:10,000 in 1% milk/PBS-T, GE Healthcare, code NA931V). Following secondary incubation, the membrane was washed three times with PBS-T and once more with PBS. For signal detection, the Immobilon Western chemiluminescent horseradish peroxidase substrate was used according to the manufacturers’ instructions. The signal was visualized on a ChemiDoc MP Imaging System (v.3.0.1.14).
Immunofluorescence analysis of γH2A expression
Immunofluorescence analysis of γH2A expression was performed as previously reported45. At least 250 cells were analysed per sample. Images were acquired with a Leica DMi8 inverted fluorescence microscope with the Leica Application Suite X (LAS X) software (v.3.7.6) and processed with Fiji (v.2.0).
FACS analysis of VSG expression
Fluorescence-activated cell sorting (FACS) analysis of VSG-2 expression was performed on live cells and therefore all steps were performed at 4 °C to prevent internalization of the VSG-2 antibody. Cells were stained immediately before analysis. For each replicate, 1.0 × 106 cells were collected by centrifugation and incubated with fluorescently conjugated anti-VSG-2 (ref. 46) diluted 1:500 in HMI-11 in the dark. Cells were washed three times with 1× TDB and resuspended in 400 µl of 1× TDB. Cells were stained with 1 µg ml−1 propidium iodide for the identification of dead cells. Samples were processed on a FACS Canto (BD Biosciences) and 10,000 events were captured per sample. Data were processed using FCS Express software (v.7). Gates were applied to remove cellular debris (FSC-A versus SSC-A) and remove doublets (FSC-A versus FSC-H).
Single-cell sorting for SL-Smart-seq3xpress library preparation
Single cells were sorted into 384-well plates for SL-Smart-seq3xpress library preparation by flow cytometry using a FACS Fusion II cell sorter (BD Biosciences) and a 100-µm nozzle within a safety cabinet. The sorter was calibrated according to the manufacturer’s protocol before collecting cells to reduce the time cells were held before sorting, thereby reducing cell death. A 384-well plate adaptor was installed and prechilled to 4 °C. Correct droplet positioning within wells was verified visually by sorting empty droplets onto a covered 384-well plate before every plate was sorted. Next, 5.0 × 106 cells were collected by centrifugation at 4 °C and washed twice in sterile filtered ice-cold 1× TDB. The cells were resuspended in 1 ml of ice-cold filtered 1× TDB, stained with 1 µg ml−1 propidium iodide and brought immediately to the sorter on ice. Populations were gated to remove cellular debris, doublets and dead cells as described above. As a consequence of our tight gating strategy (Extended Data Fig. 7), we probably enriched for cells in G1 and excluded larger cells in G2. Plates prepared with lysis buffer were thawed individually immediately before sorting and placed within the precooled adaptor. Single cells were sorted using the ‘single cell’ purity option into the appropriate wells, and the plates were immediately sealed with an aluminium foil and moved to dry ice before longer term storage at −80 °C. Sorted plates were not stored for more than 1 month before library preparation.
Generation of SL-Smart-seq3xpress RNA spike-ins
From the standard ERCC RNA spike-in set of 92 sequences, ten with a size of around 500 nt and roughly 50% G+C content were selected and synthetic spike-in DNA fragments were ordered from IDT, adding a homology region for cloning, the T7 promoter sequence and the SL sequence on the 5′ end and a homology region for cloning on the 3′ end. The fragments were cloned into a pBSIIKS+ plasmid digested with SacI and BamHI (NEB) using Infusion (Takara) and transformed into Stellar cells. Plasmids were then extracted, linearized with BamHI and in vitro transcription and polyadenylation was performed using HiScribe T7 ARCA mRNA Kit (with tailing) from NEB, following the recommended procedure. The obtained RNA from each of the ten spike-in sequences were mixed and aliquot dilutions of the spike-in mix were generated and stored at −80 °C until usage. Annotations based on the spike-in fasta file were produced with a Python (v.3.10.8) script using a biopython (v.1.81) module.
SL-Smart-seq3xpress library preparation
RNase free reagents were used for all steps and all surfaces were regularly treated with RNaseZAP (Sigma). Each well of a 384-well plate was filled with 3 µl per well silicone oil (Sigma) using an Integra Assist Plus pipetting robot. The plates were sealed with adhesive PCR plate seals and briefly centrifuged to collect the liquid at the bottom of the wells. To each well, 0.3 µl of lysis buffer (0.1% TX-100, 6.67% w/v PEG8000, 0.0417 µM oligo(dT) (5′-Biotin-AGAGACAGATTGCGCAATG[N8][T30]VN-3′), 0.67 mM of each dNTP, 0.4 U µl−1 RNase inhibitor and spike-in mix (roughly 1,364 transcripts)) were added using an I.DOT liquid dispenser (Cytena). The plates were briefly centrifuged, placed on ice and brought to the cell sorter. Single cells were sorted into each well as described above.
To lyse cells, the reaction plate was thawed, centrifuged and incubated at 72 °C for 10 min. To each well, 0.1 µl of reverse transcription mix (100 mM Tris-HCl pH 8.3, 120 mM NaCl, 10 mM MgCl2, 32 mM DTT, 0.25 U µl−1 RNase inhibitor and 8 U µl−1 Maxima H-minus Reverse Transcriptase) were immediately added following cell lysis using the I.DOT and the reaction plate then incubated at 42 °C for 90 min before inactivation of the reaction at 85 °C for 5 min. Immediately following the reverse transcription reaction, the reaction plate was centrifuged and 0.6 µl of PCR amplification mix (1.67× SeqAmp PCR buffer, 0.042 U µl−1 SeqAmp polymerase, 0.83 µM SL primer: 5′-CTAACGCTATTATTAGAACAGTTTCTGT*A*C*-3′, and 0.83 µM Reverse primer: 5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGATCATTGTAGG-3′) were added to each well using the I.DOT liquid dispenser. The PCR reaction was performed with the following conditions: 95 °C for 1 min, 16 cycles of (98 °C for 10 s, 65 °C for 30 s, 68 °C for 4 min), 72 °C for 10 min. Following the reaction, the PCR plate was centrifuged and amplified cDNA was then diluted by adding 9 µl of dH2O to each well of the plate. If not used immediately, the plate was stored at −20 °C until next step.
To each well of a new 384-well plate, 1 µl of each prediluted cDNA was added using the Integra pipetting robot. To each well, 1 µl of tagmentation mix (10 mM Tris-HCl pH 7.5, 5 mM MgCl2, 5% DMF, 0.002 µl of TDE1) was added and the plate incubated at 55 °C for 10 min. To stop the reaction, 0.5 µl of 0.2% SDS was immediately added to each well. The plate was centrifuged and incubated at room temperature for 5 min. The individual libraries generated from each well were dual indexed with Illumina i5 (5′-AATGATACGGCGACCACCGAGATCTACAC[8 bp index]TCGTCGGCAGCGTC-3′) and i7 index primers. For each index primer, 0.5 µl (2.2 µM) was dispensed into each of the reaction wells with 1.5 µl of PCR mix (3.33× Phusion HF Buffer, 0.67 mM of each dNTP, Tween-20 0.083%, and 0.033 U µl−1 Phusion HF DNA polymerase), for a final reaction volume of 5 µl. For the sequencing libraries presented in Fig. 3e (right) the volumes were the following: 0.8 µl (2.2 µM) of each index primer and 3.9 µl of PCR mix, for a final reaction volume of 8 µl. The PCR reaction was performed with the following conditions: 72 °C for 3 min, 95 °C for 30 s, 14 cycles of (95 °C for 10 s, 55 °C for 30 s, 72 °C for 1 min), 72 °C for 5 min. The reaction volumes from all wells were pooled into a robotic reservoir (Nalgene, Thermo Scientific) by centrifuging the plate placed in a custom-made three-dimensionally printed plate holder at 200g for 30 s. The pooled library was purified using AMPure XP beads at a ratio of 1:0.7. The libraries were eluted from the beads in 45 µl total volume of dH2O. To further decrease free unligated adaptor concentration, the libraries were run on a 4% non-denaturing PAGE gel and purified according to standard polyacrylamide gel purification protocols. The libraries were sequenced on a NextSeq 1000 sequencing platform to produce paired-end reads of 101 nt (cDNA read) and 19 nt (TAG + UMI read), and 8 nt for the index reads.
5′ Chromium 10X library preparation and sequencing
Cultures of N50 and P10 cells29 were set up and maintained at 0.5–1.0 × 106 cells per ml before collecting for library preparation. A mixed population sample was prepared by pooling together equal numbers of N50 and P10 cells. The mixed cells were collected by centrifugation at 400g for 10 min, washed twice in ice-cold 1× PBS supplemented with 1% d-glucose and 0.04% BSA, and resuspended in 1 ml of the buffer. The cells were then filtered with a 35 µm cell strainer (Corning) and adjusted to 1,000 cells per µl. Libraries were prepared using the Next GEM Single Cell 5′ GEM Kit v.2 (10xGenomics) and sequenced on the NextSeq 1000 platform to a depth of roughly 50,000 reads per cell. Paired-end reads of 26 nt (read 1) and 122 nt (read 2) as well as 10-nt-index reads were generated.
Primary processing of SL-Smart-seq3xpress sequencing data
The two reads containing the indexes (8 nt each) and the TAG + UMI(19 nt) were concatenated into a 35 nt read. Artefact reads containing the TAG sequence (or its reverse complement) in the cDNA read were filtered out using Cutadapt47 (v.4.3). Downsampling of reads for method benchmarking was done using seqtk (v.1.4) ‘sample’ function (https://github.com/lh3/seqtk). Reads were mapped with STARsolo48,49 (STAR v.2.7.10a) to a hybrid fasta file combining the T. brucei Lister 427 strain genome (Tb427v11, ref. 5) and the spike-in sequences, producing a transcript count matrix and an alignment (BAM) file. The count matrix was then corrected using the index hopping filtering pipeline scSwitchFilter (described in the next section) using the BAM file as input.
Index hopping filtering
scSwitchFilter (https://github.com/colomemaria/scSwitchFilter) corrects index hopping in multiplexed sequencing libraries using raw BAM files instead of a count matrix50. The correction process involves three main steps: (1) BAM to SAM conversion; (2) read extraction and parsing and (3) negative correction count matrix computation. In the first step the pipeline uses samtools51 (v.1.17) to convert a BAM file to a SAM file. In step 2, a fast bash script is used to extract and parse valid reads from the SAM file, select reads with cell barcode, UMI barcode and gene nametags, and split cell barcode tags for subsequent analysis. The selected reads are then complied into a single .TSV file. Depending on the number of plates (individual libraries) in the sequencing experiment (run), the script may split the cell barcode tag into plate–library-i5-i7 or i5-i7 barcode combinations. In step 3, scSwitchFilter calculates read counts for switched indices, assuming a low probability of the combination of UMI barcode, gene name and an index being present in several plate wells. Reads with more than 80% (default threshold) of total counts among those with switched indices remain unfiltered. The tool generates a residue count matrix that should be subtracted from the initial count matrix to obtain the filtered count matrix.
SL-Smart-seq3xpress data analysis
Count matrices were processed with JupyterLab (v.4) notebooks using IPython (v.7.31) using the following modules: pandas (v.1.5.3), numpy (v.1.23.5), scipy (v.1.10.1), scanpy (v.1.7.2), openpyxl (v.3.1.2), matplotlib (v.3.6.3) and seaborn (v.0.12.2). Cells with fewer than 500 genes detected, 1,000 gene UMI transcript counts and 50 spike-in UMI counts were filtered out. For the gene expression analysis, transcript counts for each cell were normalized by spike-in counts. For the quantification of cells expressing each VSG, we defined a cell as expressing a given VSG, if the transcript counts for that VSG represented more than 80% of the transcript counts for all VSGs in that cell. If no VSG reached this threshold, we defined the cell as having ‘no dominant VSG’. Final figures were created with Graphpad Prism (v.9).
Sensitivity and specificity comparison for different sequencing approaches
For Smart-seq2 and SL-Smart-seq3xpress data, reads were subsampled to match the average reads per cell in Chromium 10X (roughly 75,000 reads per cell in Briggs et al.26 and roughly 100,000 reads per cell for Chromium 10X data from this study). All sequencing data were mapped with STARsolo with identical settings. For the sensitivity comparison, the transcript end coordinate annotations were extended until the beginning of the next transcript, matching the conditions used by Briggs et al. using a perl (v.5.32.1) script with a perl-bioperl (v.1.7.8) module. For specificity comparison, cells with genes detected, total transcript counts or total VSG transcript counts below half of the median of the population of cells, were filtered out. Furthermore, only cells with more than ten VSG UMI counts were considered.
Type of switching analysis
Single-cell BAM files were extracted from STARsolo output BAM file, using the cell-specific cell barcode:Z attribute (storing the indexes and the TAG sequence) for each mapped read in the BAM file. Only cells with a dominant VSG from 0 h, 96 h and 10 days postinduction time points were considered. Coverage files (Bigwig files) were generated for each single cell using deepTools52 (v.3.5.4) bamCoverage function with ‘–normalizeUsing RPKM’ and ‘–minMappingQuality 10’ options. Coverage tracks were plotted using pyGenomeTracks53 (v.3.8). For the determination of switching type (recombination or transcriptional) and for the identification of the transcriptional signal end position in BES1, and the start of transcriptional signal in the BES where the newly active VSG was originally located, the single-cell coverage tracks were visually inspected in Integrative Genomics Viewer (IGV)54 (v.2.16.0).
Identification of VSG homologues
Homologues of VSG-2, VSG-8 and VSG-11 were identified by BLAST (v.2.14.0) to the Lister 427 genome assembly in TriTrypDB55. Hits with a bitscore greater than 1,000 were selected as highly similar homologues and putative ‘donors’ for segmental gene conversion. For VSG-2, VSG-8 and VSG-11, there were zero hits, five hits and one hit meeting this criterion, respectively.
Single-cell de novo VSG transcript assembly after DSB induction in VSG-8
Fastq files were demultiplexed into single-cell fastq files with deML56 (v.1.1.13) with default settings. De novo transcript assemblies were then generated for each single cell with Trinity57 (v.2.15.1), restricting the output to contigs bigger than 1 kb. To identify which of the assembled transcripts was the active VSG, the de novo assembled contigs were aligned with BLAST58 (v.2.14.0) to VSG-8, and the contigs with high similarity (bitscore greater than 2,000) were extracted using a Python (v.3.10.8) script and reheadeded the fasta sequences by cell ID using seqkit (v.2.5.1). Those cells with no contig reaching the threshold were discarded. Multifasta files with all the single-cell de novo assembled VSGs per experiment, together with the putative ‘donor’ VSGs, were constructed and aligned to VSG-8 with minimap2 (ref. 59) (v.2.10). Finally, the alignments were visualized in IGV and the start and end position of recombination and the putative donor(s) for each cell was determined.
Bulk RNA-seq library preparation and sequencing
Cell lines expressing different VSGs—VSG-2, VSG-8, VSG-11 and those used for the ATAC-seq experiments—were maintained at 0.5–1.0 × 106 cells per ml before collection. RNA-seq library preparation was performed as previously described60. Strand-specific RNA-seq library concentrations were measured in duplicate using Qubit double-stranded DNA HS Assay Kit and Agilent TapeStation system. The libraries were quantified with the KAPA Library Quantification Kit according to the manufacturer’s protocol and sequenced on the Illumina NextSeq 1000 platform to generate paired-end reads.
Bulk RNA-seq data analysis
For the bulk transcriptome analysis of Lister 427 bloodstream form wild-type cells (VSG-2 expressers) and clones that have switched to the expression of different VSGs (VSG-8 or VSG-11), reads were mapped to the Lister 427 genome assembly v11 with STAR48 (v.2.7.10a). Coverage files were generated and plotted in the same way as for the scRNA-seq data (section ‘Type of switching analysis’). For the analysis of the transcriptional switch time courses, reads were mapped with bwa-mem61 (v.0.7.17) and PCR duplicates were filtered out with Picard (v.3.2.0) ‘MarkDuplicates’ function. Counts for each gene were calculated with Subread (v.2.0.1) ‘featureCounts’ function62, filtering low confidence mapping reads (‘-Q 10’). Gene counts were then normalized to kilobases per million mapped reads.
ATAC-seq library preparation
The ATAC-seq libraries were prepared following the protocol by Müller et al.10 with several modifications. Briefly, 26.7 × 106 cells were collected (10 min at 1,800g) and washed in 30 ml of ice-cold 1× TDB. The cells were resuspended in 200 µl of permeabilization buffer (100 mM KCl, 10 mM Tris-HCl pH 8.0, 1 mM DTT, 25 mM EDTA) supplied with protease inhibitors. After adding 2 µl of 4 mM digitonin, the cells were incubated for 5 min at room temperature. Next, the cells were pelleted at 1,200g for 10 min at 4 °C, resuspended in 400 µl of isotonic buffer (100 mM KCl, 10 mM Tris-HCl pH 8.0, 10 mM CaCl2, 5% glycerol) with protease inhibitors and pelleted again. Tagmentation was performed by adding 50 µl of tagmentation mix (25 µl of 2× reaction buffer, 24 µl of dH2O, 1 µl TDE1) to the cell pellet and incubating at 37 °C for 30 min. The DNA was then purified using Qiagen MinElute PCR Purification Kit, eluted in 10 µl of elution buffer (10 mM Tris-HCl, pH 8.0), and amplified for 13 cycles using Phusion High-Fidelity DNA Polymerase with 2.5 µl of index primers (each, 25 mM) in a 50 µl of reaction mixture. The resulting libraries were purified using AMPure XP beads at a 1.8× ratio and eluted in 20 µl of nuclease-free water. The libraries were sequenced on a NextSeq 1000 platform to generate paired-end reads of 60 nt each to a depth of 400 million reads.
ATAC-seq data analysis
Reads were mapped to the Lister 427 genome assembly v.11 with bwa-mem61 (v.0.7.17). Counts per million normalized coverage per 25-nt bin was calculated with ‘bamCoverage’, whereas filtering reads with low mapping quality (‘-Q 10’). The average coverage for each BESs was calculated with ‘multibigSummary’ function from deepTools52 (v.3.5.1). For each BES and sample, the log2 ratio relative to the initial silent state was calculated.
BLISS
BLISS was performed as previously described63 (with the modifications described below). Furthermore, starting cell concentration was adjusted according to previous BLISS experiments in trypanosomes39. Cas9 was induced by incubating 2 × 108 cells with doxycycline for 4 h before cell collection. Cells were pelleted for 10 min at 800g and resuspended in 17.5 ml of warm 1× TDB, followed by fixation in 2% methanol-free formaldehyde for 10 min at room temperature with rotation. Formaldehyde was quenched by addition of glycine to a final concentration of 125 mM and incubation with rotation for 5 min at room temperature and 5 min on ice. Crosslinked cells were pelleted and washed in 20 ml of ice-cold 1× TDB, transferred to a 1.5 ml of protein LoBind tube (Eppendorf) and washed again with ice-cold 1× TDB. Crosslinked cells were counted using a Neubauer chamber and kept at 4 °C for up to two weeks before starting the BLISS template preparation. Next, 5 × 107 crosslinked cells were lysed in 200 µl of lysis buffer 1 (10 mM Tris pH 8.0, 10 mM NaCl, 1 mM EDTA, 0.2% Triton X-100) for 1 h on ice and pelleted and incubated in 200 µl of prewarmed lysis buffer 2 (10 mM Tris pH 8.0, 150 mM NaCl, 0.3% SDS) for 1 h at 37 °C with shaking at 400 rpm. The cells were washed twice with 200 µl of prewarmed CSTX buffer (1× rCutSmart buffer with 0.1% Triton X-100). DSB blunting of the sample was performed using the Quick Blunting Kit (NEB) for 1 h at 25 °C with shaking at 400 rpm and followed by two washes with 200 µl of CSTX buffer. Four microlitres of sample-specific 10 µM annealed BLISS adaptors were ligated to the blunted DSBs for 20 h at 16 °C using T4 DNA ligase (Thermo Fisher). Ligated samples were washed twice with 200 µl of CSTX buffer before resuspension in 100 µl of Tail buffer (10 mM Tris pH 7.5, 100 mM NaCl, 50 mM EDTA, 0.5% SDS) with 10 µl of proteinase K (NEB, 800 U ml−1). Samples were incubated overnight at 55 °C with shaking at 800 rpm, followed by the addition of another 10 µl of Proteinase K (NEB, 800 U ml−1) and incubation was continued for another hour. Proteinase K was deactivated by incubating the samples at 95 °C for 10 min. The template DNA was extracted using phenol–chloroform–isoamyl alcohol mixture, followed by ethanol precipitation and eluted in 130 µl of TE buffer (10 mM Tris pH 8.0, 1 mM EDTA). The extracted template DNA was sonicated in microtubes for 80 s using a Covaris S220 and the following settings: duty factor 10%, PIP 140 W, 200 cycles per burst. Sheared DNA was then purified with 0.8× AMPure XP beads and eluted in 15 µl of nuclease-free water and analysed on the TapeStation. To prepare BLISS libraries, 50–100 ng of the purified DNA were used for in vitro transcription of the template DNA using the MEGAscript T7 Transcription Kit (Thermo Fisher) and a sample incubation of 15 h at 37 °C. Next, the DNA template was degraded with Turbo DNase I (Thermo Fisher) and the amplified RNA was purified with 1× AMPure XP beads and eluted in 6 µl of nuclease-free water. The amplified RNA was analysed on the TapeStation. Afterwards, 1 µl of the 10 µM RA3 adaptor was ligated to the purified amplified RNA using T4 RNA Ligase 2 truncated (NEB), followed by reverse transcription with 2 µl of the 10 µM RTP and Superscript IV Reverse Transcriptase (Thermo Fisher). Libraries were indexed and amplified by PCR with NEBNext Ultra II Q5 (NEB) using a library-specific RPIX indexed primer and the RP1 common primer. The PCR was performed by splitting each sample into eight different PCR tubes. Double-sided clean-up was performed on the libraries with 0.45–0.75× AMPure XP beads. Final libraries were analysed on TapeStation and the library pool was quantified using the KAPA Library Quantification Kit (Roche). The libraries were sequenced with paired-end (80 cycles for read 1 and 52 cycles for read 2) on a NextSeq1000 (Illumina) sequencing platform to a depth of 40 million reads per library.
BLISS data analysis
Forward reads were trimmed to remove library barcodes and UMIs using Cutadapt47 (v.3.5), allowing up to one mismatch. The trimmed sequences were then added to their respective read names using an available Python script39. Furthermore, to avoid cross-mapping of guide RNA (gRNA)-derived sequences on the target position, reads containing the gRNA scaffold were discarded with Cutadapt. Reads were then aligned to the Lister 427 genome assembly v.11 with bwa-mem61 (v.0.7.17) with default parameters. Aligned reads were then deduplicated using the UMI on the header of the forward read with the function ‘dedup’ of umi_tools64 (v.1.1.2). Filtered alignment files from replicate experiments were merged with samtools51 (v.1.20) ‘merge’ function and the normalized coverage for the start of the forward reads, marking the DSB positions, in 10-nt bins, was calculated with bamCoverage function from deepTools52 (v.3.5.1) with the following parameters: ‘–binSize 10’, ‘–Offset 1 1’ ‘–samFlagInclude 66’ and ‘–normalizeUsing CPM’. Finally, the ratio coverage to the one in Lister 427 bloodstream form wild-type cells was calculated with ‘bigwigCompare’ function from deepTools, and plotted for specific regions with pyGenomeTracks53 (v.3.8).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.