Animal use and care
All animal experiments were performed according to the protocols approved by the Institutional Animal Care and Use Committee of Peking University. All mice were maintained in pathogen-free conditions at the Laboratory Animal Center of Peking University on a 12–12-h light–dark cycle, with a temperature of 20–25 °C and humidity of 30–70% and access to food and water ad libitum.
Single-cell isolation from mouse early embryos
To obtain pre-implantation embryos, superovulation was induced in 4-week-old C57BL/6J female mice through an intraperitoneal injection of 7.5 international units (IU) of PMSG (San-Sheng Pharmaceutical) followed by 7.5 IU of hCG (San-Sheng Pharmaceutical) 44–48 h later and then the mice were mated with 2-month-old C57BL/6J male mice. Each set of embryos at a specific stage was flushed from oviducts or uteri of pregnant female mice at the following defined time periods after hCG administration: 22–24 h (zygote), 30 h (early 2cell), 43–45 h (late 2cell), 54–56 h (4cell), 68–70 h (8cell), 78–80 h (morula) and 88–90 h (blastocysts). The embryos were maintained in M2 medium (Sigma). Germinal vesicle-stage oocytes were collected 48 h after PMSG administration.
To collect zygotes, a cumulus mass containing several zygotes surrounded by follicular cells was transferred to 1× hyaluronidase solution (Sigma) and incubated at 37 °C for a few minutes. The zygotes were then transferred to M2 medium and their zona pellucida was gently removed by treating with pre-warmed Tyrode’s acidic solution (Sigma) for several minutes. The second polar bodies of zygotes were manually removed with a very fine glass needle.
For embryos of other stages, the zona pellucida of embryos was gently removed by treating with pre-warmed Tyrode’s acidic solution (Sigma) for several minutes. The embryos were then transferred to a pre-warmed 1:3 mixture of TrypLE (Gibco) and Accutase (Gibco) and incubated at 37 °C for several minutes until the cell boundaries become apparent. The embryos were transferred to M2 medium and manually separated into single cells using a mouth pipette with an appropriate diameter needle. The dissociated embryonic cells were transferred to a pre-chilled 200 μl tube containing 10 μl cold 1% BSA–PBS and lightly fixed with 180 μl chilled methanol drop by drop. The cells were stored at −80 °C or immediately used for subsequent experiments. All tips and tubes used for cell collection were pre-rinsed with 0.1% BSA–PBS to avoid sample loss.
IVF embryo experiments
To collect embryos from IVF, oocytes were collected from C57BL/6J female mice 15 h after hCG injection. Oocytes were incubated in a 200 μl drop of HTF (M1135, Aibei) for 30 min before addition of the sperm suspension. Sperm samples were collected from C57BL/6J male mice and capacitated by placing in a 37 °C, 5% CO2 incubator for 60 min. Next, 3–5 μl of the sperm suspension taken from the edge of the sperm capacitation drop was added to the oocyte clutches (final sperm concentration of 1–5 × 105 cells per ml) and incubated for 3–4 h at 37 °C with 5% CO2. Forcefully pipetting the oocytes up and down several times in a 10 μl volume using a 200 μl pipette helped remove excess sperm. Viable fertilized oocytes were washed and transferred to a new 35 mm culture dish containing KSOM medium (M1435, Aibei). The embryos were distributed evenly throughout the culture dish and incubated at 37 °C with 5% CO2 overnight. The early 2cell and late 2cell embryos were collected at 20 and 35 h after IVF, respectively.
Mouse ES cell culture
Wild-type V6.5 mouse ES cells were cultured at 37 °C with 5% CO2 and were maintained on 0.1% gelatin-coated plates in high-glucose DMEM culture medium containing 15% fetal bovine serum (Invitrogen), 1% penicillin–streptomycin (Hyclone), 1% MEM nonessential amino acids (Cellgro), 1% Glutamax (Gibco), 1% nucleoside (Millipore), 0.1 mM 2-mercaptoethanol (Sigma) and 1,000 U ml–1 recombinant leukaemia inhibitory factor (Millipore).
Antibodies
The following antibodies were used for TACIT (catalogue and lot numbers provided after the supplier name): H3K4me1 (1:50; Abcam, ab8895, GR3369516-1); H3K4me3 (1:200; Millipore, 04-745, 3243412); H3K27ac (1:500; Diagenode, C15410196, A1723-0041D); H3K36me3 (1:200; Active Motif, 61101, 06221007); H3K27me3 (1:200; Millipore, 07-449, 3146226); H3K9me3 (1:200; Active Motif, 39161, 30220003); and H2A.Z (1:200, Abcam, ab4174, GR279096-1). Donkey anti-rabbit-Alexa 488 (1:500; Invitrogen, A32790) and donkey anti-rabbit-Alexa 555 (1:500, Invitrogen, A31572) were used as secondary antibodies. Antibodies used in immunofluorescence staining included SOX2 (1:200; Active Motif, 39843, 2226414) and CDX2 (1:200, BioGenex, MU392A-UC, MU392A0516D).
TACIT library generation and sequencing
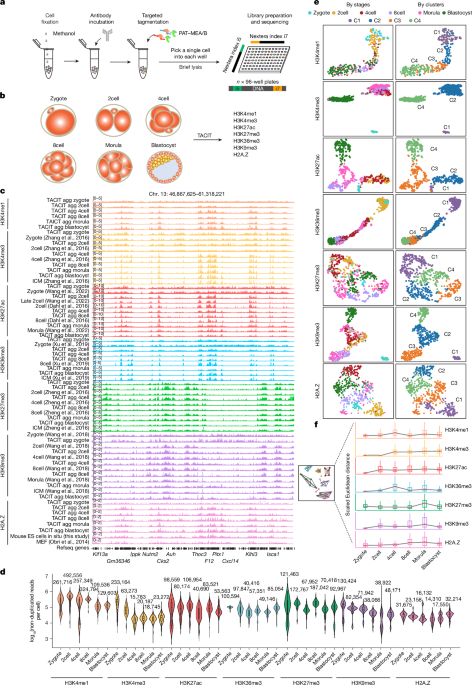
TACIT produced more non-duplicated reads than other single-cell methods for profiling histone modifications (Extended Data Fig. 1g). This improvement was attributed to the following key modifications: (1) fixing cells with methanol rather than the widely used formaldehyde; (2) tagmenting cells with the high-activity PAT enzyme as experimentally titrated; (3) reducing loss of material by titrating the incubation time for reverse-crosslinking from hours to 15 min as well as rinsing tubes and plates with 0.1% BSA–PBS; and (4) performing a single-tube reaction after pipetting into a 96-well plate for better recovery. Specifically, methanol-fixed embryonic cells or mouse ES cells were placed on ice for at least 15 min for rehydration. Cells were washed twice with wash buffer (20 mM HEPES pH 7.5, 150 mM NaCl and 0.5 mM spermidine (Sigma), 1× cocktail, 10 mM sodium butyrate and 1 mM PMSF) to remove residual methanol. We found that a brief centrifuge of cells at a low speed before aspirating the top two-thirds of the supernatant led to almost no cell loss during the washing procedure. This step ensured optimum cell recovery and satisfactory cell quality. In our experiment, different centrifugal speeds were applied to cells of different developmental stages because of the differences in the cell volume: 150g for zygotes, 200g for 2cell and 4cell stages, 350g for 8cell and morula stages and 1,000g for blastocysts.
Next, cells were incubated with specific antibody in 100 μl antibody buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM spermidine (Sigma), 2 mM EDTA, 0.01% digitonin, 0.05% TX-100, 1% BSA–PBS, 1× cocktail, 10 mM sodium butyrate and 1 mM PMSF) at 4 °C for 3–4 h. After incubation, cells were washed twice with 180 μl Dig-wash buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM spermidine (Sigma), 0.01% digitonin, 0.05% TX-100, 1× cocktail, 10 mM sodium butyrate and 1 mM PMSF) and suspended with 100 μl high-salt Dig-wash buffer (20 mM HEPES pH 7.5, 300 mM NaCl, 0.5 mM spermidine (Sigma), 0.01% digitonin, 0.05% TX-100, 1× cocktail, 10 mM sodium butyrate and 1 mM PMSF) containing 3 μg ml–1 PAT–MEA/B. The PAT expression, purification and assembly procedures were performed as per previously described guidelines19. Cells were rotated at 4 °C for 1 h to enable complete binding of PAT to antibodies and then washed twice with 180 μl high-salt Dig-wash buffer to remove free PAT–MEA/B. Tagmentation was reactivated by suspending cells with 10 μl cold reaction buffer (10 mM TAPS-NaOH pH 8.3, 5 mM MgCl2, 1× cocktail, 10 mM sodium butyrate and 1 mM PMSF) and incubated at 37 °C for 1 h in a PCR cycler. The reaction was stopped by adding 10 μl 40 mM EDTA and cells were washed twice with 1% BSA–PBS, and single cells were picked and placed into a well of a 96-well plate with a mouth pipette under a microscope. The 96-well plates were pre-rinsed with 1% BSA–PBS to avoid loss of DNA fragments, and 2 μl lysis buffer (10 mM Tris-HCl pH 8.5, 0.05% SDS and 0.1 mg ml–1 proteinase K) was added to each well. For each well, samples were covered with 5 μl mineral oil (Sigma) and incubated at 55 °C for 15 min to release DNA fragments. Next, 0.5 μl of 10 mM PMSF was added to each well to deactivate protease K, and 1 μl of 0.9% Triton X-100 was added to quench SDS in the reaction. Finally, 17 μl PCR mix (0.2 μl KAPA HiFi HotStart DNA polymerase, 4 μl 5× KAPA High-GC buffer, 0.5 μl 10 mM dNTP mix, 0.5 μl 25 mM MgCl2 and 10.8 μl H2O) was added to each well with 0.5 μl 10 mM Nextera i5 index primer and 0.5 μl 10 mM i7 index primer (Supplementary Table 1). PCR enrichment was performed in a thermal cycler with the following program: 1 cycle of 72 °C for 5 min; 1 cycle of 95 °C for 3 min; 11 cycles of 98 °C for 20 s, 65 °C for 30 s, 72 °C for 1 min; 1 cycle of 72 °C for 5 min; and hold at 4 °C. The library was purified with 1× AMPure XP beads (Beckman) once, and 200–1,000 bp fragments were selected with 0.5× + 0.5× AMPure XP beads. The libraries were sequenced with paired-end 150-bp reads on a NovaSeq 6000 platform (Illumina).
CoTACIT library generation and sequencing
For CoTACIT with embryos, isolated single cells were rehydrated and washed as described above. For the first round of barcoding, cells were incubated with 0.5 μg H3K4me3 (for the early 2cell stage) or 0.5 μg H3K27ac (for all six developmental stages) in 100 μl antibody buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM spermidine, 2 mM EDTA, 0.01% digitonin, 0.05% TX-100, 1% BSA–PBS, 1× cocktail, 10 mM sodium butyrate and 1 mM PMSF) at 4 °C for 3 h. Next, cells were washed twice with 180 μl Dig-wash buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM spermidine, 0.01% digitonin, 0.05% TX-100, 1× cocktail, 10 mM sodium butyrate and 1 mM PMSF). Cells were incubated with 3 μg ml–1 PAT-T5-1 and 3 μg ml–1 PAT-T7-1 in 100 μl high-salt Dig-wash buffer (20 mM HEPES pH 7.5, 300 mM NaCl, 0.5 mM spermidine, 0.01% digitonin, 0.05% TX-100, 1× cocktail, 10 mM sodium butyrate and 1 mM PMSF) at 4 °C for 1 h and washed twice with 180 μl high-salt Dig-wash buffer. After tagmentation and inactivation with 20 mM EDTA, cells were washed 3 times with 180 μl 1% BSA–PBS to wash out free PAT and adapters. The second round of barcoding was performed as for the first round, except that cells were incubated with 0.5 μg H3K27ac (for the early 2cell stage) or 0.5 μg H3K27me3 (for all of six developmental stages) in 100 μl antibody buffer and barcoded with 3 μg ml–1 PAT-T5-2 and 3 μg ml–1 PAT-T7-2 in 100 μl high-salt Dig-wash buffer at 4 °C for 1 h. Similarly, the third round of barcoding was carried out using the same procedure, except that cells were incubated with 0.5 μg H3K27me3 (for the early 2cell stage) or 0.5 μg H3K9me3 (for all the six developmental stages) in 100 μl antibody buffer and barcoded with 3 μg ml–1 PAT-T5-3 and 3 μg ml–1 PAT-T7-3 in 100 μl high-salt Dig-wash buffer at 4 °C for 1 h. Finally, cells were washed 3 times with 1% BSA–PBS and single cells were picked and placed into a well of a prepared 96-well plate followed by fragment release, proteinase K inactivation and SDS quenching, as described for the TACIT procedure.
Two-round PCR was performed as previously described19, which resulted in the standard Illumina Truseq Compatible library. In brief, 20 μl PCR mix (0.2 μl KAPA HiFi HotStart DNA polymerase, 4 μl 5× KAPA high-GC buffer, 0.5 μl 10 mM dNTP Mix, 0.5 μl 25 mM MgCl2, 11.6 μl H2O and 0.5 μl 50 µM in total first-round primer mix) was added to each well. PCR was performed as follows: 1 cycle of 72 °C for 5 min; 1 cycle of 95 °C for 3 min; 8 cycles of 98 °C for 20 s, 65 °C for 30 s, 72 °C for 1 min; 1 cycle of 72 °C for 5 min; and hold at 4 °C. Excess primers were digested by adding 0.25 μl ExoI (NEB) and plates were incubated at 37 °C for 60 min followed by 72 °C for 20 min. A volume of 10 μl second-round PCR mix (0.1 μl KAPA HiFi HotStart DNA polymerase, 2 μl 5× KAPA High-GC buffer, 0.25 μl 25 mM MgCl2 and 6.4 μl ddH2O) was added to each well containing 0.5 μl of 10 mM Truseq index i5 and 0.5 μl Truseq index i7 (Supplementary Table 1) and subjected to PCR with the following program: 1 cycle of 95 °C for 3 min; 5 cycles of 98 °C for 20 s, 65 °C for 30 s, 72 °C for 1 min; 1 cycle of 72 °C for 5 min; and hold at 4 °C. The library was purified with 1× AMPure XP beads (Beckman) once and 200–1,000 bp fragments were selected with 0.5× + 0.5× AMPure XP beads. The libraries were sequenced with paired-end 150-bp reads on a NovaSeq 6000 platform (Illumina).
Embryo-barcoded TACIT
For embryo-barcoded TACIT with cells of early and late 2cell stages, the zona pellucida of embryos was gently removed by treating with pre-warmed Tyrode’s acidic solution (Sigma) for 30 s. The embryos were transferred to M2 medium and directly fixed with methanol as described above. The embryos were stored at −20 °C or immediately used. Whole embryos were directly applied to the regular TACIT pipeline as described above. After tagmentation, the two cells from the same embryo were separated with custom microdissection needles and deposited into different wells of lysis buffer. Each well was covered with 5 μl mineral oil (Sigma) and incubated at 55 °C for 15 min to release DNA fragments. Next, 0.5 μl of 10 mM PMSF was added to each well to deactivate protease K, and 1 μl of 0.9% Triton X-100 was added to quench SDS in the reaction. PCR amplification was conducted as described for TACIT libraries, and DNA fragments of the two cells from the same 2cell embryos were barcoded with different combinations of Nextera i5 and i7 indexes. Finally, the embryo-barcoded TACIT library was sequenced as described for conventional TACIT.
Low-input itChIP
The itChIP–seq20 protocol was performed with a few modifications. First, the zona pellucida of embryos was gently removed by treating with pre-warmed Tyrode’s acidic solution (Sigma) for several minutes. Subsequently, embryos were transferred to M2 medium and fixed with 1% formaldehyde solution at room temperature for 3 min followed by 1× PBS wash and centrifugation at 4 °C. Samples were preserved at −80 °C or used immediately. Fixed embryos were incubated in hypotonic buffer (20 mM HEPES pH 7.9, 10 mM KCl, 10% glycerol, 0.2% NP-40 and 0.05% SDS) at 37 °C for 30 min to release chromatin. Embryos underwent gentle fragmentation by sonication (Q800R sonicator, 20% power, 10 s) and quenched with Triton X-100. Genomic tagmentation was obtained by incubating with Tn5 assembled with MEA/B adapters at 37 °C for 1 h. After the tagmentation reaction, samples were further processed to release chromatin from nuclei. After centrifugation at 4 °C, the soluble supernatant was isolated and incubated with antibodies overnight. Dynabeads protein A (Invitrogen) beads were used to pull down chromatin–antibody complexes. DNA fragments were eluted from beads and treated with proteinase K. The resultant DNA was purified and extracted using phenol–chloroform, followed by library preparation using the KAPA HiFi HotStart technique as per the TACIT procedure and supplemented with Illumina Nextra index primers. After size selection for fragments ranging from 200 to 1,000 bp, the libraries were quantified using Qubit to determine their concentration. The pooled samples were sequenced on a NovaSeq 6000 (Illumina) for paired-end 150 bp reads.
Microinjection in zygotes for siRNA knockdown
For siRNA knockdown, isolated zygotes were microinjected with sets of three siRNAs against targets (20 μM in total) or with non-target control (NC, 20 μM in total). The following siRNAs were used: NC, UGGGACUUGCAGGCCUGAUAUTT; Nanog, CGAGAACUAUUCUUGCUUATT, CCUGAGCUAUAAGCAGGUUAATT and UGGAGUAUCCCAGCAUCCAUUTT; Zfx, GGUUCAUGAUAGUGUAGUATT, GGAUGAAGAUGGACUUGAATT and GGAGGACAACGAAAUGAAATT; Yy2, GCUGCGAGAAGAUGUUCAATT, CACCAUGUGGGACGAUGUUAATT and GACCUAUAGCAUGCUCUCAUATT; Tcf12, GUGGCAGUCAUCCUUAGUCUATT, GAUGCAAUGUCCUUCUUAATT and GGAACAAGUGGUCAACCAATT; Cebpb, GAGCGACGAGUACAAGAUGTT, CACCCUGCGGAACUUGUUCAATT and CGCCUUUAGACCCAUGGAAGUTT; Bbx, UGGGACUUGCAGGCCUGAUAUTT, CCAGUGGGAGCAAGAAGUUUATT and CUCCCUCAAUAUAGUCCUAUUTT; Smad2, GUGAUAGUGCAAUCUUUGUTT, UGGUGUUCAAUCGCAUACUAUTT and CCUUCAGUGCGAUGCUCAATT; Hbp1, CCCUACCCAAUCUGCCAUAUATT, GGCUAACAGAGUUAGCAAATT and CCAGCUAAGUUCAGAUGUATT; Cdx2, GGACAGAAGAUGAGUGGAATT, GAGAAGGAGUUUCACUUUATT and GCUUGCUGCAGACGCUCAATT; Klf6, GCUUGCUGCAGACGCUCAATT, GACCAAUAGCCUGAACUCUTT and GAUGAGUUGACCAGACACUTT; Sox15, CCUGGCAGUUACACCUCUUCTT, GAUGAAGAGAAGCGACCCUUTT and GACUCUUCCACUCCAUAUAAUTT; Med1, UAAGCUUGUGCGUCAAGUAAUTT, GGCUCUCCAAUCCUUAGAACATTand GUGGCCUAUAACACUCUAAUUTT; Elf5, GCCCUGAGAUACUACUAUAAATT, GGACCGAUCUGUUCAGCAATT and GGAGGUUAGUGUACAAAUUTT; and Hif1A, CCAUGUGACCAUGAGGAAATT, GCAGACCCAGUUACAGAAATT and GCAGGAAUUGGAACAUUAUTT. siRNAs were ordered from Hippobio. The injected embryos were transferred to KSOMaa medium (Millipore) and droplets were covered with mineral oil (Sigma) in a Petri dish (Ibidi) and cultured in a tissue incubator (37 °C and 5% CO2) (Thermo Fisher Scientific). Embryos were collected at the 8cell or blastocyst stage, and single-embryo RNA-seq or immunofluorescence staining was performed to confirm KD or marker gene expression.
scRNA-seq or single-embryo RNA-seq library generation and sequencing
scRNA-seq and single-embryo RNA-seq library preparation were performed using a modified Smart-seq3 protocol51,52. The zona pellucida was gently removed by treating with Tyrode’s solution (Sigma). Isolated single cells or single embryos at the 8cell or blastocyst stage after siRNA microinjection were mouth-pipetted into lysis buffer. Lysis buffer consisted of 0.15% Triton X-100 (VWR Life Science), 5% PEG8000 (Thermo Fisher Scientific), 0,5 μM oligo-dT (Supplementary Table 1), 0.5 mM dNTPs and 0.5 U RNase inhibitor (Takara). After dispensing, lysis tubes were briefly centrifuged to ensure that lysis buffer was located under the overlay. Tubes of sorted cells were denatured at 72 °C for 10 min, followed by the addition of the reverse transcription mix. The reagent concentrations were as follows: 25 mM Tris-HCl pH 8.3 (Sigma), 30 mM NaCl (Sigma), 0.5 mM GTP (Thermo Fisher Scientific), 2.5 mM MgCl2 (Sigma), 8 mM DTT ((Thermo Fisher Scientific), 0.25 U μl–1 RNase inhibitor, 2 μM TSO (5′-AAGCAGTGGTATCAACGCAGAGTACATG(r)G(r)G(+)-3′) and 2 U μl–1 Maxima H Minus reverse transcriptase (Thermo Fisher Scientific). Reverse transcription and template switching were carried out at 42 °C for 90 min followed by 10 cycles of 50 °C for 2 min and 42 °C for 2 min. The reaction was terminated by incubating at 85 °C for 5 min. Indicated volumes of PCR master mix were dispensed, which contained 1× KAPA HiFi PCR buffer (Roche), 0.3 mM dNTPs each (Roche), 0.5 mM MgCl2 (Roche), 0.6 mM P2 primer (5′-GTGACTGGAGTTCAGACGTGTGCTCTTCCGATC-3′) and 0.2 μM IS primer (5′-AAGCAGTGGTATCAACGCAGAGT-3′) and 0.02 U μl−1 KAPA HiFi DNA polymerase (Roche). Pre-amplification was performed as follows: 3 min at 98 °C for initial denaturation, 16 cycles of 20 s at 98 °C, 30 s at 65°C, and 5 min at 72 °C. Final elongation was performed for 5 min at 72 °C. After PCR, samples were pooled and purified using a TIANquick Mini Purification kit (Tiangen) and 0.8× AMPure XP beads (Beckmann).
After purification, 1 μl cDNA was used for measuring the concentration. About 10 ng cDNA was subjected to tagmentation with 1 μM PAT–MEA in the reaction buffer (10 mM TAPS-NaOH pH 8.3 (Sigma), 5 mM MgCl2 (Sigma), 10% N,N-dimethylformamide (DMF) (Sigma)) at 55 °C for 10 min. Samples were then treated with 0.025% SDS at 55 °C for 10 min and 0.15% Triton X-100 at 37 °C for 10 min. Enrichment PCR was performed as follows: 3 min at 95 °C for initial denaturation, 16 cycles of 20 s at 98 °C, 15 s at 67 °C, and 1 min at 72 °C, and 5 min at 72 °C. The library was purified with 1× AMPure XP beads. Size selection was carried out first with 0.5× AMPure XP beads and second with 0.5× XP AMPure beads in the supernatant to obtain 200–1,000 bp fragments for sequencing. The libraries were sequenced with paired-end 150-bp reads on NovaSeq 6000 platform (Illumina).
Immunofluorescence staining
Injected embryos were fixed with 4% paraformaldehyde (Sigma) for 10–15 min. PBST (PBS + 0.5% Triton-X) was added for 20 min at room temperature to permeabilize the embryos and the samples were subsequently incubated with blocking buffer (PBS + 0.1% Tween-20 + 5% NDS) for 4 h at 4 °C. After blocking, embryos were incubated with SOX2 (Active motif) and CDX2 (BioGenex), diluted in blocking buffer, at 4 °C overnight. Samples were then washed with PBS 3 times and incubated with secondary antibodies (Invitrogen), diluted in blocking buffer, for 2–4 h at room temperature. Finally, blastocysts were incubated with 600 nM DAPI solution (Thermo Fisher) for 5 min at room temperature and were rinsed with PBS before visualization. Images were acquired using a confocal microscope (Zeiss LSM 710).
CRISPRa-mediated TF activation in mouse ES cells
The sgRNAs targeting the promoter of each of the candidate totipotency-related TFs were synthesized and inserted into a CROP-opti vector separately (Addgene, 106280) (Supplementary Table 9). Three libraries of sgRNAs included candidate TFs (CEBPG, LBX1, ETS2, MEF2D, ESR2, ESR1 and ALX1), positive control TFs (ZSCAN4 and DUX) and a non-targeting control as previously described53 at equal molar ratios. The supernatant with lentivirus was collected 18 h after transfection and filtered to remove cell debris. The mouse ES cells were infected (8 μg ml–1 polybrene) with various titres of lentivirus to achieve different multiplicity of infection values. At 24 h after transduction, new culture medium with 2 μg ml–1 puromycin was added for 48 h for selection. Cells after transduction and selection were collected for scRNA-seq. Cell pellets were fixed with 1% formaldehyde at 4 °C for 10 min and were preserved at −80 °C or used immediately. Single-cell RNA-seq for mouse ES cells for capturing both mRNA and sgRNAs was conducted as per the SPLiT-seq pipeline as previously described54.
Data processing
TACIT data were processed as previously described19, but with a few modifications for single cells. Raw TACIT sequencing data were evaluated using FastQC (v.0.11.5), followed by mapping to the mouse reference genome mm10 by Bowtie2 (v.2.2.9)55. Mapped reads with MAPQ vales less than 30 were considered as multi-mapped reads and filtered out using Samtools (v.1.9). PCR duplicates were also removed using Picard (v.2.2.4). For aggregated analysis, single-cell .bam files were merged with Samtools. For peak calling, MACS2 (v.2.1.1)56 with the ‘–broad’ parameter was used to call peaks for aggregated profiles of TACIT data. Raw CoTACIT sequencing data were de-multiplexed and paired using an in-house code as previously described19. Sequencing data for each histone modification was performed according to the analysis pipeline as described for TACIT data.
Correlation analysis for TACIT data
For correlation analysis between different experiments, we calculated the normalized mean scores in 5-kb bins of the genome by using the multiBigwigSummary function in deepTools (v.3.5.1)57. The Spearman correlation or Pearson correlation was calculated between replicates and plotted using the plotCorrelation function.
Genome-coverage analysis
To calculate the genome coverage at each developmental stage, we first called peaks for aggregated .bam files of each histone modification. We used MACS2 to call peaks with parameters of ‘–nolambda–nomodel -q 0.05–broad’. Next, we binned the mm10 genome into 200-bp genomic intervals, and for each histone modification, genome coverage at a specific stage was calculated as the percentage of genome intervals that overlapped with peaks at that stage. To evaluate genome coverage for single cells, the genome was first binned into 200 bp and bins with histone modification signals ≥ 1 were defined as covered bins. The percentage of covered bins was defined as genome coverage for each single cell.
Clustering of TACIT and CoTACIT data
TACIT alignment files were converted to a matrix with genomic intervals (instead of peaks) as rows and cells as columns using cisTopic (v.0.3.0)58. For different histone modifications, different sizes of genomic intervals were used as follows: 5 kb bins for H3K27ac; 10 kb bins for H3K4me3, H3K36me3 and H3K27me3; and 15 kb bins for H3K4me1 and H3K9me3. Clustering of embryonic cells on the basis of histone modifications was performed using Seurat (v.4.3.0). In brief, the cell–bins matrix was first normalized with the term frequency–inverse document frequency (TF–IDF), followed by dimensionality reduction with singular value decomposition (SVD). Next, 2:20 or 1:20 (only for H3K4me3) dimensions were used for identifying clusters and for UMAP visualization. For clustering of CoTACIT data from the early 2cell stage, fragment counts in 5 kb genome windows were used for all three histone modifications. The Seurat (v.4) WNN59 framework was used to generate a multimodal representation using dimensions 1:20 (H3K4me3), 2:20 (H3K27ac) and 2:20 (H3K27me3).
Normalization of Euclidean distance
To evaluate cell heterogeneity among stages for each histone modification, we first calculated the Euclidean distance between each pair of cells in the same stage as shown in the UMAP embeddings. The median Euclidean distance of zygotes was set as the baseline for normalization of other cells across all stages.
Generation of synthetic cells
To investigate the dynamics of chromatin states during mouse pre-implantation development, we generated synthetic cells as follows:
-
1)
We ordered scRNA-seq cells along the developmental trajectory using Monocle3 (ref. 60) and merged five adjacent single cells along pseudotime into one RNA synthetic cell.
-
2)
We integrated H3K4me1, H3K4me3, H3K36me3 and H3K27ac TACIT profiles with gene expression. In brief, the cell–peak or cell–bin matrix for each histone modification was first generated using cisTopic58. The GeneActivity function of Seurat (v.4) was used to create a gene-activity score matrix based on the cell–peak or cell–bin matrix. Next, anchors between the two modalities were identified with the FindTransferAnchors function. In particular, many titrations were performed to obtain the highest prediction score, including using the cell–peak or cell–bin matrix, or the bin size of the cell–bin matrix. TACIT cells with a prediction score lower than 0.2 were filtered out. Notably, for integrating cells in the 2cell stage, histone-modification signals in non-canonical broad binding regions were excluded before Seurat integration (5 kb for H3K4me3 and H3K27ac, 20 kb for H3K36me3 and H3K4me1).
-
3)
We integrated H3K27ac CoTACIT profiles with gene expression in the same way as described in step (2). As H3K27ac, H3K27me3 and H3K9me3 profiles were experimentally linked, we directly transferred corresponding H3K27me3 and H3K9me3 profiles to the linked RNA synthetic cells.
-
4)
Having obtained 155 RNA synthetic cells interpolated with six histone-modification profiles, we performed hierarchical clustering with RNA synthetic cells on the basis of multimodal histone modifications. The number of clusters closely corresponded to the exact cell number of each developmental stage, such as two clusters for the 2cell stage, four clusters for the 4cell stage, and so on. Next, we aggregated histone-modification profiles of cells in the same cluster, which led to 90 synthetic single cells with joint profiles of 6 histone modifications. To reduce effects from sequencing depth, we normalized cell numbers and non-duplicated reads before aggregating data.
WNN analysis for interpolated single cells
After integrating the TACIT and CoTACIT data with the RNA data, we obtained interpolated single cells simultaneously with six histone-modification measurements. We used the Seurat (v.4) WNN framework to generate a multimodal representation of interpolated single cells. We used the FindMultiModalNeighbors function to generate a WNN graph using the following dimensions: H3K4me1, 2:15; H3K4me3, 2:15; H3K27ac, 2:15; H3K36me3, 2:15; H3K27me3, 2:20; and H3K9me3, 2:15.
ChromHMM for synthetic cells
To integrate the six histone modification profiles, we used the multivariate HMM introduced in ChromHMM61. We binarized all .bam files for each synthetic cell using the binarizeBam function of ChromHMM with default parameters. We used the LearnModel function with default parameters to learn 12 states separately on each synthetic cell. To reduce noise and mitochondrial interference, all reads from mitochondrial DNA are filtered out. Next, we annotated each state in three steps: (1) filtering out chromatin states with extremely low genome coverage (<0.001%), because these were probably from technical noise; (2) defining hidden chromatin states based on the combination of histone modifications; and (3) correcting the annotation on the basis of the overlap in the main genome categories. Finally, we labelled the 12 states as multivalent (all histone modifications), weak promoters (H3K4me3), strong promoters (H3K4me3 and H3K27ac), weak enhancers (H3K4me1), strong enhancers (H3K4me1 and H3K27ac), poised gene bodies (H3K36me3 and repressive histone modifications), active gene bodies (H3K36me3 and active histone modifications), polycomb-protein-associated heterochromatin (only H3K27me3), H3K9me3-associated heterochromatin (only H3K9me3), heterochromatin (H3K27me3 and H3K9me3), and quiescent/low.
scChromHMM for interpolated cells
To annotate chromatin states at single-cell resolution for blastocyst cells, we first generated single-cell profiles with simultaneous measurements of six histone modifications. As described above, we integrated H3K4me1, H3K4me3, H3K27ac and H3K36me3 TACIT profiles as well as H3K27ac CoTACIT profiles with gene expression. In addition, we annotated ICM and TE cells on the basis of the expression of ICM or TE marker genes.
We used the LearnModel command of ChromHMM46 to train a 12-state model with aggregate ICM and TE profiles. Next, we ran the forward–backward algorithm to learn the posterior probability distribution for interpolated single cells. We set up the bin size of 2,000 bp and grouped the states into 6 categories (enhancers, promoters, gene bodies, polycomb-protein-associated heterochromatin, H3K9me3-associated heterochromatin and quiescent/low). We also merged five adjacent single cells along pseudotime52.
Clustering by scChromHMM-defined chromatin-state annotations of synthetic single cells
For clustering cells on the basis of scChromHMM-defined chromatin-state annotations of all genomic intervals (Fig. 4a), we used the posterior probability matrix for each state as input for TF–IDF normalization, SVD dimensionality reduction, cluster finding and UMAP visualization with Seurat (v.4). We used 1:5 dimensions for clustering and visualization. For clustering cells on the basis of chromatin-states annotations in all TSSs, we selected genomic intervals that were ±2 kb flanking TSS regions and averaged the posterior probability for a specific chromatin state in each TSS as the average probability of this chromatin state. Next, we used the mean probability matrix for each chromatin state for TF–IDF normalization, SVD dimensionality reduction, cluster finding and UMAP visualization. Dimensions 1:5 were used for clustering and visualization.
Identification of differential bins between totipotent and pluripotent cells
In Extended Data Fig. 10b, for each chromatin-state labelling, after TF–IDF normalization, SVD dimensionality reduction and cluster finding (as described above), we used the FindMakers function in Seurat (v.4) to find differential bins between cells at the 2cell and 8cell stage. Next, to quantify the establishment of totipotency-related chromatin regions during development, we calculated the percentage of differential bins that were already annotated as corresponding chromatin states for each synthetic cell.
Identification of feature classifier bins for defining totipotency
We adopted two strategies for identifying putative totipotency-related classifier bins between 2cell 1 versus 2cell 2 (strategy 1) and 2cell versus 8cell (strategy 2), in which the former aimed to rule out differences in stage. For each strategy, a cell–bin probability matrix was first generated for each chromatin state, and genomic regions for which posterior probability exhibited a correlation of more than 80% or less than −50% with the expression of totipotency marker genes was selected. Next, all highly correlated candidate bins were aggregated to generate the state matrix with state annotation information for each genomic interval. The state matrix of synthetic cells at the 2cell and 8cell stages was used as input for constructing the random forest training model62, with labels as ‘toti-high’ and ‘toti-no’ groups, respectively.
Identification of feature classifier bins for ICM and TE specification
With the scChromHMM annotation for blastocyst synthetic cells, we used 29 synthetic cells as training cells to build a random forest machine-learning model and 10 synthetic cells as testing cells for cross-validation. This trained random forest model prioritized 780 genomic intervals for which chromatin states are essential for the first lineage specification and predicted the ICM or TE tendency of 4cell, 8cell and morula cells.
TF motif analysis
In Extended Data Fig. 7g, we intersected our ChromHMM-annotated regions with ATAC-seq peaks and called motifs on these ChromHMM–ATAC regions using Homer. By default, Homer uses random genomic regions as the background. For motif analysis in Figs. 4e and 5e, we directly used classifier bins for calling motifs because the bin size was 200 bp and 2,000 bp, respectively. We considered that such bins are sufficiently narrow for calling TF motifs. To disentangle the influence of open chromatin during TF motif enrichment assessment in classifier bins, previously published ATAC-seq data (GSE66390)18 were used. All open-chromatin regions from the 2cell and 8cell stages were combined and used as the background for Fig. 4e. ATAC-seq peaks were segmented into 200-bp bins, matching the classifier bin size. First, 765 TF motifs were identified on classifier bins that were annotated as active states (promoter, enhancer and gene bodies) for all synthetic cells. Next, we selected 327 TF motifs that were highly enriched in 2cell 1 or 2cell 2 (–log10(P) > 8) and depleted in 8cell (–log10(P) < 2). Finally, only TF motifs with detected expression in 2cell 1 or 2cell 2 (TPM > 2) were chosen as putative totipotency-related TFs, as listed in Supplementary Table 8. In Fig. 5e, TF motif enrichment was calculated using Homer for classifier bins that were annotated as enhancers for each synthetic cell. Open-chromatin regions from the 4cell to blastocyst stages were used as the background, with ATAC-seq peaks binned to 2,000 bp, matching the classifier bin size. Only synthetic cells defined as ICM-potential or TE-potential were used for TF motif enrichment analysis during ICM or TE specification, respectively. We enriched 59 TF motifs on classifier bins that were annotated as active states for ICM-potential cells and 42 for TE-potential cells (–log10(P) > 5). Next, we selected eight putative ICM-related TFs and five putative TE-related TFs on the basis of expression levels (RPKM > 1.5 in Ribo-lite data63).
Distribution of classifier bins
The enrichment of the classifier bins in Extended Data Fig. 10c was calculated using observed versus expected probability as previously described12. The observed probability was calculated using the length of classifier bins covering the related genomic regions versus the length of the total classifier bins, and the expected probability was calculated using the length of the total related genomic regions versus the length of the mouse genome. Promoter was defined as ±1 kb genomic region around all TSSs. The locations of annotated repeats (RepeatMasker) were downloaded from the UCSC Genome browser18,64.
Gene expression with chromatin states
For each synthetic cell, the median gene expression was presented from cells belonging to the same synthetic cell. Each chromatin region was linked to the nearest genes using Homer, and expression for all genes and all samples were then combined and split by categories of chromatin states. A boxplot was plotted for each chromatin state. To eliminate the effects of non-canonical chromatin-binding features, only synthetic cells at the 2cell and 4cell stages were included in this analysis.
Identifying promoter–enhancer pairs
To identify promoter–enhancer pairs, we used TSS-proximal signals (± 5 kb flanking TSSs) to build a peak–cell matrix for H3K4me3 and used TSS-distal signals to build a peak–cell matrix for H3K27ac. Next, we integrated these two matrices and evaluated co-occurrence of pairs with Cicero (v.1.14.0)65. We also established a criterion in which only pairs that link a H3K4me3 peak and a H3K27ac peak were defined as promoter–enhancer pairs. Overall, we identified 43,983 promoter–enhancer pairs (Cicero score, S > 0.1). To find putative functional promoter–enhancer pairs implicated in ZGA, we chose pairs for which H3K4me3 peaks fell within ±2 kb flanking the TSS of genes that are activated after ZGA66. We calculated the fold change (FC) of the Cicero score between 2cell 1 and 2cell 2 clusters (FC = S2cell 2/S2cell 1). We defined 2cell 1-specific (FC < 0.25, S2cell 1 > 0.3, and S2cell 2 < 0.05), 2cell 2-specific (FC > 2, S2cell 1 < 0.05, and S2cell 2 > 0.3) and shared (S2cell 1 > 0.3, and S2cell 2 > 0.3) promoter–enhancer pairs. Finally, we identified 515 2cell 1-specific, 1,138 2cell 2-specific and 159 shared promoter–enhancer pairs.
Similarly, we chose promoter–enhancer pairs for which H3K4me3 peaks or H3K27ac peaks fell into copies of MERVL elements as MERVL-associated pairs, whereby MERVLs function as promoters or enhancers, respectively. We identified 332 enhancer–promoter pairs in which MERVL elements functioned as promoters, and 866 enhancer–promoter pairs in which MERVL elements functioned as enhancers.
Re-analysis of Hi-C data
The allvalidPairs matrix for the late 2cell stage was downloaded from the GEO database (accession number GSE82185)67. To identify interactions, we used the analyzeHiC function of Homer (v.4.11)68 at 50 kb resolution and plotted interactions with Python (v.3.9.7).
Chromatin states and expression of transposable elements
For the enrichment analysis in Fig. 4f, the observed probability was calculated using the length of classifier bins covering the related transposable elements versus the length of the total classifier bins, and the expected probability was calculated using the length of the total related transposable elements versus the length of the mouse genome. Overall, we identified 75 transposable elements that were highly enriched (log2(overexpression) > 1) in 2,583 totipotency-related classifier bins. For the annotation of chromatin states of the enriched 75 transposable elements in Fig. 4g, we calculated the percentage of transposable element copies that were defined as promoters, enhancers, gene bodies, heterochromatin or quiescent/low regions for each synthetic cell. To quantify expression levels of these transposable elements, we mapped raw scRNA-seq reads to the mm10 genome using Hisat2 (v.2.2.1)69 and filtered out mapped reads with MAPQ less than 10 with Samtools (v.1.9). We calculated the numbers of TPM based on the locations of annotated repeats (RepeatMasker) downloaded from the UCSC Genome browser.
Analyses for multiplexability of CRISPRa in mouse ES cells
To evaluate the efficacy of totipotency activation of our CRISPRa experiments in mouse ES cells, we first quantified the abundance of designed sgRNAs targeting candidate TFs (CEBPG, LBX1, ETS2, MEF2D, ESR2, ESR1 and ALX1), positive control TFs (ZSCAN4 and DUX), and non-targeting control based on detected sgRNA unique molecular identifiers (UMIs). sgRNAs with fewer than 16 UMIs were filtered out for further analyses. Perturbed cells were clustered and projected in UMAP together with totipotent blastomere-like cells and pluripotent stem cells in public scRNA-seq datasets42 using Seurat (v.4). To analyse the perturbation effects of candidate TF genes, we ranked the genes and calculated perturbation correlation among of them based on cells receiving only one sgRNA using MUSIC70 with default parameters. Furthermore, the totipotency score for cells with each combination of gene perturbation was calculated on the basis of the totipotent gene signature.
RNA-seq analysis for single embryos with siRNA knockdown
Sequenced reads with adaptor and low-quality bases were removed. The clean reads were aligned to the mm10 reference genome (RNA library) and in-house siRNA database (siRNA library) with Hisat2. For each single embryo, the expression level of a gene was normalized by the TPM. The KD information for each embryo at the 8cell and blastocyst stage as metadata was incorporated together with the gene-expression matrix in Seurat analysis. The individual embryos were visualized by UMAP using Seurat (v.4) with default parameters. The TF activity for cells with TE or ICM candidate gene KD was evaluated using SCENIC71.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.