
The robotics laboratory at Insilico Medicine, an artificial-intelligence biotechnology firm in Boston, Massachusetts.Credit: Insilico Medicine
Drug discovery is extraordinarily difficult. “In 100 years or so of contemporary medicine, we’ve found treatments for only around 500 of the roughly 7,000 rare diseases,” says David Pardoe, a computational chemist at Evotec, a biotechnology company in Hamburg, Germany. “It takes too long and costs too much.” But in theory, and to the excitement of many, artificial intelligence (AI) could address both of these problems.
AI should be able to bring together the 3D geometry and atomic structure of a potential drug-like molecule, and build a picture of how it fits into its target protein. Designs can then be tweaked to make a potential drug more potent, or an algorithm might identify whole new targets to pursue. An AI system might also take into account the essential backdrop to interactions between drugs and their target: the complex biological milieu of a patient’s body. Unwanted interactions with various non-target proteins might burden an otherwise promising molecule with side effects.
The key to developing systems that are capable of boosting the drug-discovery process is lots of good data. Compared with scientists in some other fields in which AI is being deployed, researchers who seek to apply the technology to drug development have a solid foundation on which to build: large volumes of biological data are being produced in laboratories all over the world, all the time.
Nature Outlook: Robotics and artificial intelligence
But, although the scale of the available data might indicate that the AI transformation of drug development is surely just a matter of time, this will not necessarily be the case. The quality of the data, most of which are not collected with machine learning in mind, is not always up to scratch. Lack of consistency in experimental methods and how data are recorded can pose problems, as can a bias towards publishing only positive results. Although some people think that a greater volume of data will be a solution in its own right, others think that academic and industry researchers will have to come together to improve the quality of the data that are fed to machine-learning models.
Which problems are most pressing, and which solutions should be prioritized, are subject to debate. Nature spoke to a number of researchers who are active in this field, in an effort to tease out some of the actions that could be taken to enable AI to transform drug discovery and development to the extent that many people hope it will.
Standardize reporting and methods
Academic scientists pride themselves on their flexibility. Experiments are run with the equipment they have. If an opportunity to incorporate a better machine or process into their methods becomes available, they will take it. This attitude keeps them on the frontiers, but it might pose a problem for machine learning. “A huge issue for AI is how data are generated,” says Eric Durand, chief data science officer at AI biotech firm Owkin in Paris.
When different laboratories use different methods, reagents and machines, discrepancies known as batch effects can be introduced into the resulting data. Slight differences in the protocols used to process samples, variation between batches of reagents and cells and even something as basic as how a molecular structure is described or named, are sources of variation that a pattern-hungry AI model might incorrectly interpret as biologically meaningful. “You can’t just take data sets that were generated by two labs and co-analyse them without preprocessing,” says Durand.
This undermines the utility of many large public databases launched before the AI tidal wave. ChEMBL, for example, is a free, curated database of bioactive molecules that pools information from studies, patents and other databases, and is widely used in drug discovery. It is maintained by the European Bioinformatics Institute based in Hinxton, UK, and tries to minimize batch effects, but the way in which it aggregates information means that inconsistencies still arise. “You need to be careful,” says Pat Walters, a computational chemist at Relay Therapeutics, a biotech company in Cambridge, Massachusetts. “You have data from labs that didn’t do experiments in the same way, so it is difficult to make apples-to-apples comparisons.”
The best approach to generating the ordered data that AI requires, some argue, would be to lay down rules of how to run and report experiments. The names of diseases and genes can be harmonized from the start, for instance, and protocols agreed ahead of time. One example of this in action is the Human Cell Atlas, a global project launched in 2016 that has already mapped millions of cells in the human body in a rigorous and standardized way. This provides consistent data, which are ideal fodder for AI algorithms that are searching for potential drug targets.
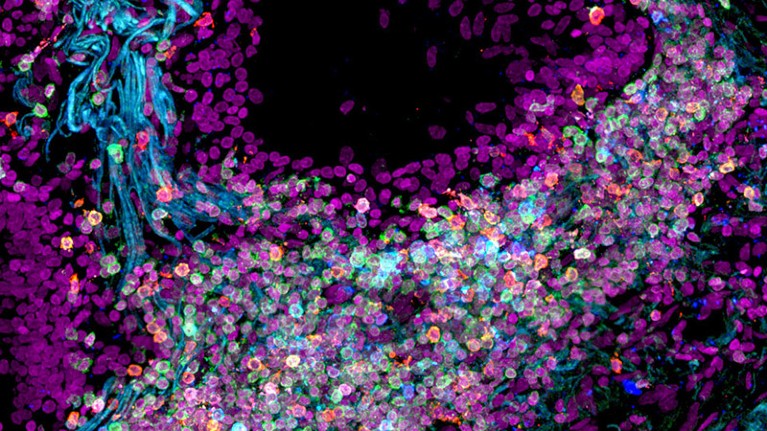
Human lung cells have been mapped by the Human Cell Atlas, a global consortium that aims to create comprehensive reference maps of all human cells.Credit: Nathan Richoz Univ. Cambridge
An endeavour called Polaris — a benchmarking platform for drug discovery — also aims to help clean up and standardize data sets for machine learning. It set out guidelines in a preprint article1 in late 2024 and is now looking for feedback. Polaris outlines basic checks for data sets. For instance, creators must explain how data are generated and should be used, and they must reference sources that they have drawn from. It warns that it is up to the creator to check for obvious duplicates and ambiguous data. “We also get experts to vet some publicly available data sets to let the community know which are of high quality and which are not,” says Walters. Polaris has introduced a certification stamp for those that cut the mustard.
Without further effort to generate harmonized and relevant data, there might be little value in continuing to develop ever more advanced algorithms, says Pardoe. “Once those ‘good’ data are available, then we can make rapid and significant progress in the right direction.”
Recognize the value of negative results
For academic investigators, there is often little to gain from reporting that your experiment failed. Those who try typically find it difficult to get their research published. This built-in bias towards positive results in science is nothing new, but it poses a particular problem for the use of AI.
Data taken from published work and fed into an algorithm will invariably present a distorted, rose-tinted view of the biological landscape. Because there are more data available on mature compounds that have shown promise in animals with no toxic effects than data on unsuccessful compounds, for example, an AI model for drug discovery will be mostly deprived of information on the many hidden failures.
Miraz Rahman, a medicinal chemist at King’s College London, points to an example of such bias in the pursuit of new antibiotics. A crucial step in killing microbes is to sneak a compound inside the bacterial cell, and many published studies suggest that primary amines — small compounds similar in structure to ammonia — help to get drugs inside bacteria. “If you asked an AI model, based on published studies, it would keep suggesting compounds containing primary amines,” Rahman says. But he knows he would have to ignore this advice. “My lab has got so much data showing that this doesn’t work,” he says. The problem for AI is that failures such as Rahman’s have not been published.

Medicinal chemist Miraz Rahman says that publishing only positive results causes problems for artificial-intelligence tools.Credit: King’s College London
The same bias towards sharing positive results also afflicts pharmaceutical companies. “What is being published? Always the success story,” says Rahman. When companies choose not to make their negative findings public, the facade presented to algorithms is simpler and shinier than reality.
One way around this is to start out with the express intention of compiling both negative and positive results. One such project that is gaining attention is led by James Fraser, a structural biologist at the University of California, San Francisco, and funded by the US Advanced Research Projects Agency for Health. Its interest lies in what is called pharmacokinetics, which describe what the human body does to a compound.
A drug’s fate depends on how it is absorbed, distributed, metabolized and excreted by the body — known collectively as ADME. If the body gets rid of a compound too slowly, it might pose safety risks. But it might also be ejected too fast. “You can come up with a molecule that binds very tightly to your target of interest, but if that molecule is rapidly excreted, it has no value as a drug,” says Walters, who is an admirer of the project. Drugs can also interact with non-target proteins in the body, which might trigger toxicities and slow down or reduce the amount of drug that reaches the desired destination.
These ADME issues tend to surface only late in a drug’s development, and can lead to expensive failures. “The process is kind of whack-a-mole right now,” says Fraser. “You make new molecules to get rid of a liability, and then another liability pops up and you again optimize around that.”

James Fraser is a structural biologist at the University of California, San Francisco.Credit: Deanne Fitzmaurice/UCSF
Fraser refers to his current work, which aims to provide AI tools with the data they need to spot these pitfalls, as the “avoid-ome” project2, because it seeks to generate data not on drug targets, but on the proteins that researchers normally want to avoid. The intention is to build a library of experimental and structural data sets on protein binding that is relevant to ADME. Since funding was approved in October, his lab has begun by running tests on metabolic aspects of ADME.
The results should help to create predictive AI models that can optimize the pharmacokinetics of drug candidates. “People will make fewer molecules, with a better holistic view of all potential liabilities, and get to a molecule that passes all criteria and gets to humans faster,” says Fraser.
Share industry data and expertise
Drug companies keep large amounts of data, including negative results, and strive to collect them in a standard way that makes it ideal for AI models to digest. Only a small proportion of these data, however, make it into the public realm — Rahman estimates that the more-open pharmaceutical companies publish only about 15–30% of their data, increasing to up to 50% for clinical trials.
The value of the data they hold is not lost on the companies themselves. In 2018, Vas Narasimhan,chief executive of the pharmaceutical company Novartis in Basel, Switzerland, described reimagining the organization as a “medicines and data science company”, stressing its ambition to embrace AI in drug discovery. As a result, most pharmaceutical firms are deeply averse to sharing data with academics and with each other.
“A company like Novartis — which I worked in for many years — has tens of thousands of compounds that they assessed for binding to certain proteins,” says Durand. “But they don’t want to share that with competitors, because it is really a main asset.”

Eric Durand is the chief data science officer at the artificial-intelligence biotechnology firm Owkin in Paris.Credit: Owkin
One effort to encourage data sharing between pharmaceutical companies, in which Owkin was involved, was an EU-funded project called Melloddy. The project employed a federated learning approach, which allowed ten companies to collaborate in training predictive software without revealing sensitive biological and chemical data to their competitors3. During this project, models that were trained to relate a molecule’s chemical structure to its biological activity were significantly more accurate than most of the companies’ existing models.
Not everyone was smitten, however. Familiar problems that arise when combining data that have been produced in different ways also apply to data from different companies. Moreover, the anonymization of the data might reduce their richness. “It’s very difficult to combine data sets without revealing the chemical structures and the nature of the assays that were used to generate them,” explains Walters.
The project also did nothing to improve the state of the public data sets that are relied on by academics, who are smart enough to know that no amount of asking is going to let them waltz through drug companies’ data libraries. Instead, some researchers are calling on pharmaceutical companies to apply their other big advantage to public databases: money.
One of the most valuable data sets available to researchers is the UK Biobank, which has systematically collected genetic, lifestyle and health information, as well as biological samples, from 500,000 people in the United Kingdom. The effort has received more than £500 million (US$632 million) in funding — but mostly from government and charitable sources. “Big pharma companies should be sponsoring initiatives such as UK Biobank,” says Alex Zhavoronkov, founder and chief executive of Insilico Medicine, an AI biotech firm based in Boston, Massachusetts.
Do more with what you have
Some researchers argue that sheer volume of data — and smarter processing — will go a long way to overcoming the difficulties of using AI for drug discovery. “With enough data you can learn how to generalize,” says Zhavoronkov.
Insilico Medicine links data that are produced as a result of billions of dollars in US government research grants to publications, clinical trials, patents and repositories of genetic and chemical data. “Modern AI tools allow you to trace the innovation back to the grant,” Zhavoronkov says.

Alex Zhavoronkov (left), founder and chief executive of Insilico Medicine, says that pharmaceutical companies should support projects such as the UK Biobank, a biomedical database that contains information from 500,000 people. Credit: Insilico Medicine
This raw material is then processed. At Insilico, this involves introducing scores that, for example, help the algorithms to weigh the importance or veracity of results. “We’ve got a tool which evaluates the credibility of the scientist that publishes a paper,” says Zhavoronkov. “If a person has lied before, then the probability of them lying again is higher.” Insilico also tracks the movement of stock prices after companies announce clinical-trial results. If the price dropped precipitously, it is assumed the results were negative, no matter what a firm says.
In late 2019, Insilico’s AI-driven drug-discovery platform PandaOmics discovered a target for fibrotic diseases that involve excessive scar tissue. It then used its generative AI platform Chemistry42 to find compounds that would block this target4. The algorithm taps into large databases of existing molecules, including ChEMBL, to learn patterns in chemical structures and to design potential drugs. In August last year, Insilico completed a phase IIa study of a small molecule for use in adults with a lung-scarring disease called idiopathic pulmonary fibrosis. The company is now plotting a follow-up trial, and Zhavoronkov expects more success to come. “Since 2019 we’ve nominated 22 preclinical candidates,” he says.
Some large public data pools will undoubtedly be harder for AI to make use of. Consider bulk RNA sequences, for example. Compiled from mashed-up tissue samples, they average the output of genes across many cells. Today, the sequences of individual cells are viewed as superior, because they can be used to detect what proteins rare cells are making and they give better resolution when mapping a tissue. But Zhavoronkov advocates against ignoring any less-than-ideal public data. “That must be reused. A lot of animals were sacrificed, and AI needs to be trained on those data,” he says. He advocates for the creation of smaller, high-quality data sets that can be used to test whether an AI model that has been trained on larger, potentially flawed data sets is making good predictions. These data can come from automated labs, he suggests, set up to generate specific types of data in a standardized way.



