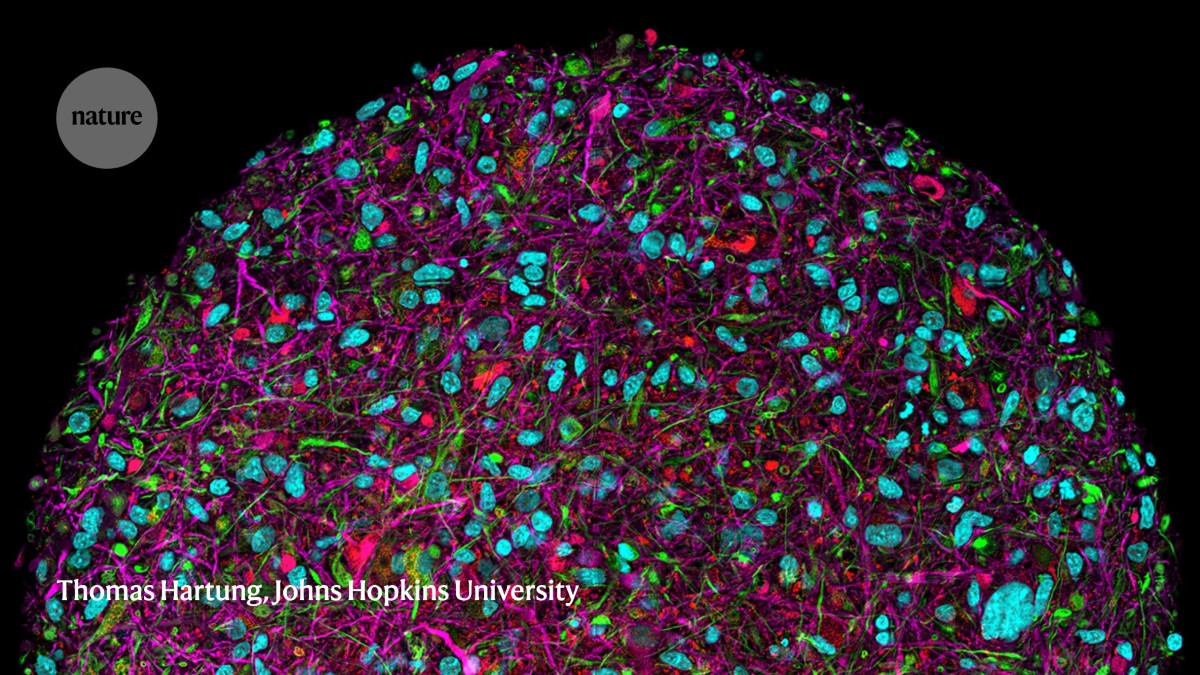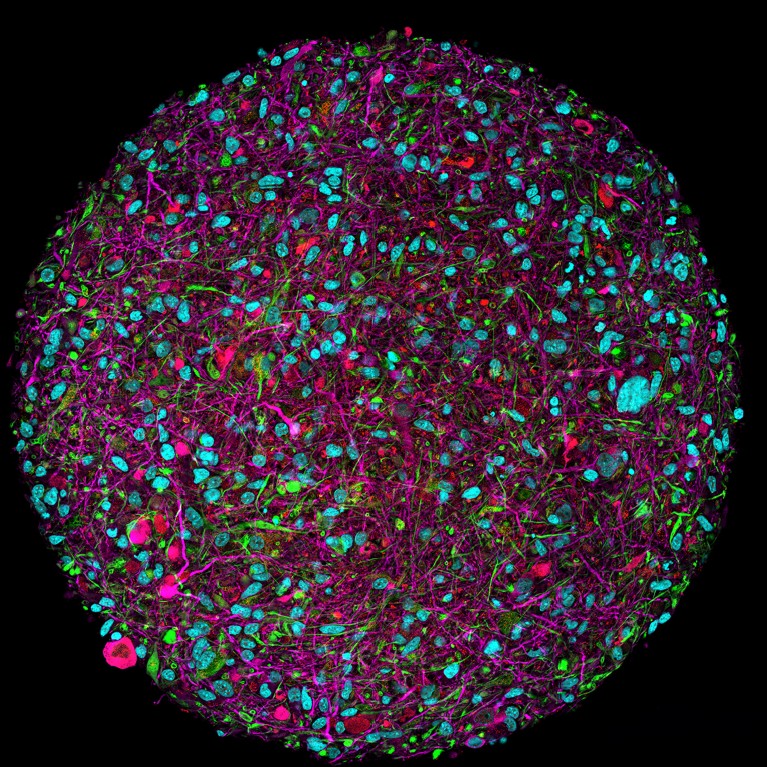
A magnified image of a brain organoid with fluorescent labelling of various cell types.Credit: Thomas Hartung, Johns Hopkins University
Advanced tools and techniques are giving researchers a detailed look at the body’s defence system. From spotting single cells in action to improving or replacing animal models, the methods described below are helping immunologists to push the boundaries of their work in 2025.
THX mice: mouse on the outside, human on the inside
Since the first mouse models with human cells and tissues were developed more than 40 years ago, they’ve become much better at mimicking the human immune system. But they’re still nowhere near accurate enough.
One major issue is that mice have more than 1,600 immune-response genes that do not match their human equivalents. And most ‘humanized’ mice (which have human genes, cells or tissues) don’t mount enough antibodies to fight infections, which is a problem for researchers who study vaccines and immune responses, says Paolo Casali, an immunologist at the University of Texas Health Science Center at San Antonio. “They are not useful at all,” he says.
Nature Index 2025 Immunology
A team led by Casali and his former graduate student Daniel Chupp, now an immunologist at Massachusetts biotechnology company Invivyd, has been trying to overcome this hurdle for decades, and their efforts are beginning to pay off. In a paper published last year1, the team introduced THX, a mouse model that replicates the human immune system.
To create THX, mice are injected with human stem cells that develop into immune components such as lymph nodes, antibodies and T and B cells (types of white blood cells). When given an mRNA-based COVID-19 vaccine, they mounted a strong immune response — an important trait for vaccine research.
THX mice have caught the eye of several research groups in the United States and Europe. “I get e-mails almost every day,” says Casali. Among those groups is one led by James Voss, an immunologist at Scripps Research in La Jolla, California, who plans to use the mice to advance his work on targeted HIV therapies. Voss and his team have used CRISPR gene-editing to reprogram human B cells to produce antibodies that can recognize and block many different strains of HIV. And although existing humanized mouse models have B cells, they fail to mount a strong enough antibody response, creating what Voss calls a “developmental block”. THX mice overcome this limitation, opening a new path for his research.
Once they reach the market, THX mice are expected to be relatively cheap and less labour-intensive to produce than other mouse models. This is because they don’t require complicated tissue engraftments, says Voss, who hopes to start using them in the coming months. “I’m very excited. I think this will be a real game-changer.”
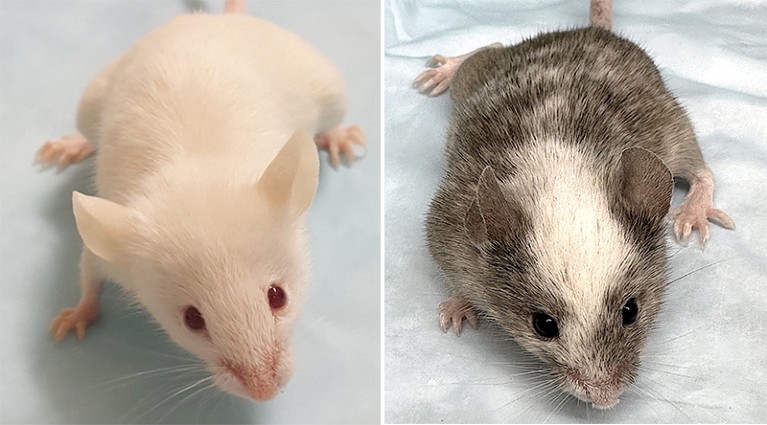
Different variants of the THX mouse model, which replicates the human immune system.Credit: Daniel Chupp (left); Carlos E. Rivera (Right)
Organ-on-chip and organoid systems: move over, mice
Some researchers are doing away with animal models altogether by creating miniature versions of human organs and tissues. These systems, which include organ-on-chip platforms and organoid systems (cell cultures that mimic organ functions), are providing snapshots of the human gut, liver, lung and other organs in both sickness and health.
It’s a tantalizing prospect for drug development, because these systems could provide a more accurate model of human biology than animal models, says Thomas Hartung, a pharmacologist at Johns Hopkins University in Baltimore, Maryland. He adds that many drugs that have been approved over the past decade are based on human proteins and antibodies. “It makes no sense to test them in animal models.”
Tiny organ models are also helping researchers to better understand how systems in the body interact with each other and respond to disease. Linda Griffith, a bioengineer at the Massachusetts Institute of Technology in Cambridge, and her colleagues have developed a model of the human gut to study how the organ’s microbes interact with immune cells and regulate inflammation. They have also created models for endometriosis and pancreatic cancer.
Building systems like these — and trying to reproduce them — is difficult, and not something that many labs are able to do, says Griffith. She adds that scaling them up for mass production is also very complicated, because it requires intensive quality-control measures.
Despite these hurdles, organ and tissue microsystems are likely to become more common in labs, at least in the United States. In July, the US National Institutes of Health (NIH) announced that it will no longer offer new funding to projects that rely solely on animal models to study human disease. Instead, the NIH will prioritize studies that use human-based technologies, including organ-on-chip and organoid systems.
The US Food and Drug Administration (FDA) announced similar plans this year to phase out animal testing in favour of human cell cultures and organoids for drug safety trials. The FDA’s roadmap noted that animal models have been poor predictors of drug success for many common diseases, from cancer to Alzheimer’s. Some compounds that seemed safe in animal models have been lethal in humans, according to the regulator. “They’re telling us very clearly this is what the future looks like,” says Hartung.
ScRNA-seq: levelling up single-cell sequencing
Few techniques have helped researchers to unravel the mysteries of the immune system quite like single-cell RNA-sequencing (scRNA-seq) — a method to determine which genes are being transcribed into the RNA of individual cells. Introduced in the late 2000s, scRNA-seq has rapidly evolved over the past decade, and has been used to map specific immune cell types and to pinpoint key differences in gene activity that affect immune response. Now, researchers are matching scRNA-seq with other tools to give it different capabilities.
One method that pairs particularly well with scRNA-seq is CITE-seq, which identifies both RNA and proteins on the surfaces of individual cells simultaneously, says Sara Suliman, an immunologist at the University of California, San Francisco.
This combined approach has been a boon for research on natural killer (NK) cells (white blood cells that destroy diseased cells), says Eric Vivier, an immunologist at the Centre for Immunology of Marseille-Luminy in France. Last year, Vivier’s team published a study2 that used scRNA-seq and CITE-seq to uncover three main types of NK cell based on the proteins they produce in various tissues and cancers. This helped the researchers to design better cell ‘engagers’ — molecules that guide NK cells to attack tumours. The team went on to build a new engager that activates all three NK cell types to target treatment-resistant forms of non-Hodgkin’s lymphoma3.
Such efforts would not have been possible using older technologies, such as bulk RNA sequencing, which obscures the distinct signatures of individual cells, says Vivier.
Another scRNA-seq add-on, called single-cell ATAC-seq, highlights which DNA regions are open, accessible and potentially active in regulating gene expression, rather than simply showing which genes are switched on. The technique not only offers insights into cellular function, but also allows researchers to piece together what’s possible or primed to happen in the future. In August, a team led by researchers at the Tsinghua University School of Medicine in Beijing described how they used ATAC-seq to study acute myeloid leukaemia, a rapidly progressing cancer of the blood and bone marrow4. The work showed how faulty gene regulation in some leukaemia cells can cause the disease to return after treatment.
Perturb-seq, meanwhile, is a technique that combines scRNA-seq with CRISPR gene-editing. This allows scientists to switch off or modify genes across thousands of cells simultaneously, and to track how those changes affect the cells. It’s been used to study cause-and-effect relationships, such as how a small genetic change might make someone more vulnerable to infection or inflammation, says Suliman. “That is where the powerful data is.”