Generation of m11 chimeric Fc protein and m11-specific antibodies
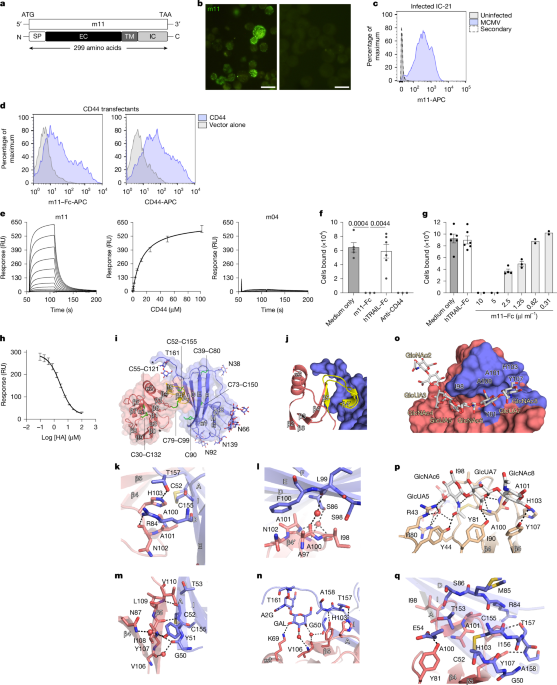
The MCMV m11–Fc (also referred to as vCD44BP–Fc) fusion protein was generated using Sew-PCR to attach the Fc portion of human IgG1 to the part of the m11 gene that encodes its extracellular domain (amino acids 1–212). The construct was transiently transfected into CV-1/EBNA cells and the soluble fusion protein purified using protein-A-Sepharose (Amersham Pharmacia Biotech). A human TRAIL–Fc fusion protein was used as a negative control. A monoclonal antibody (M-627) to m11 was generated by immunizing mice with the m11–Fc protein and fusing splenocytes with a myeloma partner using standard methodologies. The specificity of M-627 was verified using m11 transfected cells and by comparing cells infected with wild-type MCMV and a mutant lacking m11, referred to as ΔvCD44BP—see below. The anti-m11 monoclonal antibody (7G5) was generated by immunizing rats with the m11 ectodomain (amino acids 28–164) and fusing splenocytes with X63 myeloma cells using standard methodologies. Supernatants from hybridoma clones were initially screened by ELISA using plate bound m11–Fc protein and a secondary screen performed by flow cytometry using fibroblasts infected with either MCMV or ΔvCD44BP virus.
Expression cloning of the m11 cognate from a CD40 ligand-stimulated B cell cDNA library
A cDNA library was generated from mouse B cells stimulated with CD40 ligand and cloned into the pDC409 vector using previously described methods53. Approximately 200 pools, each containing ~2,000 clones, were transfected into CV-1/EBNA cells and 2 days later the transfected cells screened using the m11–Fc protein as described54. One positive pool was identified, and this was subdivided into smaller pools until a single positive cDNA was obtained from the original pool. Individual clones were then sequenced, and sequences were compared to public DNA databases.
Construction and cloning of m11 for transfection
The MCMV m11 open reading frame (ORF) was amplified by PCR based on published sequences (accession number AM886412) using purified MCMV DNA templates. The m11 PCR product was cloned into the pDC409 mammalian expression vector54 to generate p409-m11.
Cell lines
COS-7 and EL4 cells were cultured in Dulbecco’s modified Eagle medium (DMEM, Gibco); IC-21 cells were cultured in Roswell Park Memorial Institute (RPMI-1640, Gibco); M2-10B4 and mouse embryonic fibroblasts (MEFs) were cultured in minimal essential medium (MEM, Gibco). All culture media were supplemented with 10% fetal calf serum (FCS) (Gibco) (COS-7, EL4 and IC-21) or 10% newborn calf serum (NCS) (M2-10B4 and MEF), and antibiotics (penicillin 100 µg ml−1, CSL; gentamycin 40 µg ml−1, Pharmacia & Upjohn).
The fibroblastic reticular cell line (FRC2) was generated by A.L.F. by isolating FRCs from the lymph nodes of C57BL/6J mice as described55 and FRC enriched via CD45 and CD31 depletion (Miltenyi Biotec). Stromal cells were plated overnight in alpha-MEM with 10% FBS (Gibco) and then co-transfected using Lipofectamine 3000 with a piggyBac expression vector expressing SV40LT with GFP and a piggyBac transposase expression vector, hyPBase (The Sanger Centre, pCM-hyPBase).
CD44–hyaluronic acid binding assay
The hyaluronic acid adhesion assay was performed with modifications of a previously described method56. In brief, 96-well plates (Costar) were coated with 100 µl of a 100 µg ml−1 solution of hyaluronic acid (220 kDa) and incubated overnight at 37 °C. The plates were then gently washed and incubated with 200 µl of 2% BSA in PBS for 2 h at 37 °C. Separately, 2 × 105 EL4 cells previously treated for 24 h with PMA (100 ng ml−1) and ionomycin (500 ng ml−1) (Thermo Fisher Scientific) were added to the wells of 96-well plates containing 10 µg ml−1 of anti-CD44 antibody (KM114, that masks the hyaluronic acid binding site), m11–Fc or the irrelevant human TRAIL–Fc, and incubated for 30 min at 4 °C. An anti-human-Fc antibody (10 µg ml−1, Jackson ImmunoResearch) was then added to the wells containing the Fc proteins and the plates incubated for a further 30 min at 4 °C. These cells were then transferred to the hyaluronic acid-coated plates in DMEM containing 10% FCS, and incubated for 1 h at 37 °C. After three washes with PBS, 100 µl of DMEM were added back into the wells. To quantify the number of cells in the wells, an MTT colorimetric assay (Sigma-Aldrich, St Louis, MO, USA) was used and the absorbance read at 570 nm on a AD200 microplate reader (Beckman-Coulter Inc.). MTT incorporation levels reflect number of cells present in the wells as determined microscopically with the number of EL4 cells calculated using a standard curve.
Protein chemistry
The construct encoding mouse CD44 (amino acids 23–174) was cloned into the p30 vector and expressed as inclusion bodies in TonA-BL-21 Escherichia coli cells. CD44 was refolded by dilution in a solution containing 4 M urea, 0.4 M l-arginine, 0.1 M EDTA, 0.1 M Tris-HCl pH 8.0 in a 5:1 mM reduced:oxidized glutathione overnight at 4 °C. Refolded CD44 was purified first via DEAE anion exchange and size-exclusion chromatography using a Superdex S75 16/600 column (GE Healthcare). The m11 ectodomain (amino acids 28–164 from MCMV strain K181) was cloned into the pFASTBac vector (Invitrogen) to include a C-terminal hexa-histidine tag. The plasmid was incorporated into a recombinant baculovirus, and the viral titre expanded in SF9 cells as described in the Bac-to-Bac manual (Invitrogen). Soluble m11 was obtain by infecting Hi5 cells with 2% P3 virus. The construct encoding m04 (amino acids 24–223 from MCMV strain G4) was cloned into the pHLSec vector to include a C-terminal hexa-histidine tag and expressed via transient transfection in human embryonic kidney 293-S cells as described13. Secreted proteins from mammalian and baculoviral systems were buffer-exchanged into 10 mM Tris-HCl (pH 8.0), 500 mM NaCl and purified via nickel-affinity and size-exclusion chromatography using a Superdex S200 16/600 column (GE Healthcare) in a 10 mM Tris-HCl (pH 8.0), 150 mM NaCl buffer.
Crystallization and data collection
An equimolar mixture of m11 and CD44 was resolved using a Superdex S200 16/600 column (GE Healthcare) and pure heterodimer was concentrated to 9.65 mg ml−1 in 20 mM Tris-HCl pH 8.0 and 150 mM NaCl. Crystals were obtained using the hanging drop vapour diffusion method from a mother liquor containing 0.2 M LiSO4, 0.1 M Tris-HCl pH 8.5 and 30% (w/v) PEK 3000. Prior to data collection, crystals were cryoprotected in mother liquor supplemented with 10% (v/v) glycerol. Crystals were flash cooled using liquid nitrogen and X-ray diffraction data was recorded using a Quantum-315 CCD detector at the MX2 beamline of the Australian Synchrotron. Data were integrated by MOSFLM and scaled using SCALA within the CCP4 suite of programmes (Extended Data Table 1).
Structure determination and refinement
The structure was determined by molecular replacement using Phaser. Isolated models for CD44 and m11 were generated from the structure of mouse CD44 (PDB ID: 2JCP) and m04 (PDB ID: 4PN6), respectively, using PyMOL (Schrödinger, Inc.). The structure was refined via iterative cycles of model building in Coot and refinement using Buster (http://globalphasing.com/buster/). N- and O-linked glycans were manually incorporated into regions of positive density that correlated to the requisite sequence motif: NX(S/T), where X is any amino acid except proline, serine or threonine. The final structure was refined to final Rfactor/Rfree values of 18.4% and 20.2%, respectively. Details of the refinement statistics are provided in Extended Data Table 1. The structure factor file and associated atomic coordinates have been deposited in the PDB under accession code 9EJW.
Surface plasmon resonance
SPR experiments were performed using a BIAcore 3000 system (GE Healthcare) at 25 °C with a buffer comprising 10 mM Tris-HCl pH 8.0, 300 mM NaCl, and 0.005% (v/v) surfactant P20. CM5 sensor chips (GE Healthcare) were primed with an equal mixture of EDC (1-ethyl-3-(3-dimethylaminopropyl)carbodiimide) and NHS (N-hydroxysuccinimide). The m11 and m04 ectodomains were prepared in buffer containing 150 mM NaCl and either 20 mM sodium acetate pH 5.3 or 100 mM sodium citrate pH 3.0, respectively, and approximately 2,300 response units were immobilized per flow cell. Flow cells were quenched with 20 μl of ethanolamine at a flow rate of 5 μl min−1 and primed twice with running buffer prior to injection of analyte. Varying concentrations of CD44 or hyaluronic acid (100–0.1 μM) pre-incubated with a fixed concentration (12.5 μM) of CD44 were passed over the flow cells for 65 s, in duplicate, at a flow rate of 10 μl min−1. The final responses were double referenced by subtracting responses from an ‘empty’ flow cell. The responses at equilibrium were used to construct equilibrium binding curves that fit by a single-site binding model. The calculated equilibrium dissociation constants represent the mean ± s.e.m. from two independent experiments. Data were analysed with Scrubber (BioLogic Software) and Prism (GraphPad Software).
Generation of the ∆vCD44BP virus and revertant virus
A homologous recombination approach was used to generate the m11 mutant virus. A 5 kb Hpa1 fragment spanning residues 7114–12176 (Genbank accession AM886412) of the K181 strain of MCMV was cloned into pBluescript SK−. The 4.2 kb LacZ cassette from the MV10 vector was subcloned into the Nde1 site at nucleotide positions (10,948–10,953) within the m11 ORF. Following linearization, the plasmid was co-transfected into MEFs together with purified K181 MCMV DNA. Plaques were screened for β-galactosidase expression using X-gal staining and β-gal+ plaques were plaque purified to generate a ∆m11 stock. To introduce a premature stop codon into the m11 ORF, Sew-PCR was used to generate a PCR product in which the unique Nde1 site in m11 was replaced with an Hpa1 restriction site encompassing an in-frame premature stop codon. This construct was used to co-transfect MEFs with purified ∆m11 viral DNA. Viral preparations in which the LacZ cassette has been substituted by the Hpa1-containing construct were selected by identification of plaques that did not stain blue with X-gal staining. Plaques were purified to homogeneity and a single clone selected and designated ∆vCD44BP. An m11 revertant virus (REV) was generated by co-transfection of ∆m11 with plasmid constructs containing wild-type K181 sequence spanning the m11 region. All mutants were sequence verified across the m11 region and restriction fragment length polymorphism analysis was performed to compare the profiles of the stop mutants, revertant and wild-type viruses.
Mice
BALB/c and C57BL/6J mice were purchased from the Animal Resources Centre/Ozgene ARC (Perth, Western Australia, Australia) or the Walter and Eliza Hall Institute of Medical Research (Melbourne, Victoria, Australia). B6 Cd44−/− (ref. 57) and B6 BALB-TC1 (TC1) (H2b NK1.1 + Ly49H–)38 mice were bred at Perkins Bioresources Facility (Perth, Western Australia, Australia). BALB/c.Ifng−/− mice and BALB/c.Prf1−/− mice were obtained from the Animal Services Facility at QIMR Berghofer Medical Research Institute (Queensland, Australia). Age-matched adult female mice (8–12 weeks old) were used as controls for all experiments. All animal experimentation was performed with ethics approval from Monash University Ethics Committee (MARP2); Perkins Animal Ethics Committees (for the Lions Eye Institute); University of Western Australia Animal Ethics Committee (for the Lions Eye Institute) and in accordance with NHMRC Australia Code of Practice for the Care and Use of Animals for Scientific Purposes.
Viral infections and in vivo monoclonal antibody administration
Mice were infected intraperitoneally with MCMV (K181 strain), ∆vCD44BP, MCMV-REV or MCMV-K181-Perth-mCherry salivary gland-propagated virus (5 × 103 or 1 × 104 PFU, except for BALB/c. Prf1−/− which were infected with 2 × 103 PFU owing their increased susceptibility to infection) diluted in PBS containing 0.05% FCS. For CD8 or CD4 T cell depletion studies, mice were injected intraperitoneally with anti-CD8β monoclonal antibody (clone 53.5.8, 250 µg per injection) or anti-CD4 monoclonal antibody (clone GK1.5, 500 µg per injection) at days −2, 0 and 2 relative to virus injection. Depletion of CD4 or CD8 T cells was confirmed by flow cytometric analysis.
For influenza infections, mice were infected intraperitoneally with 1.5 × 107 PFU of the influenza A virus (IAV) strain A/Puerto Rico/8/34 (H1N1, PR8) diluted in PBS.
For intravenous antibody labelling of dendritic cells, 1.5 µg of anti-CD11c APC antibody (clone HL3) was injected 36 h post-infection and mice were humanely killed three minutes later. Spleens were collected and single-cell suspensions were prepared using the stromal isolation protocol described in ref. 55, stained with the appropriate antibodies, and analysed by flow cytometry.
Generation of bone marrow chimeras
Bone marrow cells were collected from the tibia, femur and ilium of TC1 donor mice and washed with sterile PBS. Recipient WT or B6 Cd44−/− mice received two doses of 500 cGy total-body irradiation, spaced 3 h apart, prior to receiving 107 bone marrow cells administered by intravenous injection. Chimeric mice were housed for three months to allow full reconstitution of the haematopoietic compartment. Chimerism in the haematopoietic compartment was >95% in this system.
CRISPR-mediated deletion of CD44
Bone marrow cells were isolated from TC1 mice and haematopoietic stem cells were enriched using an EasySep Mouse Hematopoietic Progenitor Cell Isolation Kit following the manufacturer’s instructions (STEMCELL Technologies, 19856 A). The purified progenitor cells were seeded into 24 wells plates at 1 × 106 cells per well and incubated at 37 °C for 2 h in 1.5 ml of growth medium consisting of StemSpan SFEM II medium (STEMCELL Technologies, 09605) supplemented with 50 ng ml−1 Stem cell factor (SCF) (Thermo Fisher Scientific, PMC2113L). CD44 deletion was achieved by precomplexing two Cd44 single guide RNA (sgRNA) guides (300 pmoles of Cd44 sgRNA1 plus 300 pmoles of Cd44 sgRNA2) with 36.3 pmoles of Cas9 protein (IDT, 1081059). Each reaction was in a total of 5 μl and was incubated at room temperature for 10 min. In addition, control electroporation reactions using a scrambled non-targeting sgRNA were performed. P3 Nucleofector Solution (Lonza, PBP3-00675) was prepared by mixing 16.4 μl of P3 Solution with 3.6 μl of Supplement 1 and the solution was allowed to equilibrate to room temperature. Progenitor cells were washed once in PBS before being resuspended in P3 Nucleofector Solution (5 × 105 cells in 20 μl) and the cells added to the guide–Cas9 mixture. A total of 20 μl of the cell plus guide–Cas9 mixture was transferred to a well of a 16-well Nucleocuvette strip and pulsed using the unstimulated mouse T cell programme (4D-Nucleofector X Unit, Lonza). Multiple electroporations were performed to generate a sufficient number of cells. Immediately after electroporation, 80 µl of warm growth medium was added to each well and the strip incubated at 37 °C for 30 min. Cells were then transferred to a 6-well plate and cultured in growth medium at 37 °C for a further 2 days. Cells from individual cultures were pooled and loss of CD44 confirmed by flow cytometry before being injected into lethally irradiated C57BL/6J (CD45.1) recipient mice (2 × 105 cells per mouse). Details of the sgRNA guides are provided in Extended Data Table 2.
Quantification of viral loads
Viral titres were quantified as described58. In brief, individual organs were homogenized in MEM containing 2% NCS and the homogenate was centrifuged at 3,000g for 15 min at 4 °C. The supernatant was collected, and viral titres determined by adding serial dilutions of the supernatant to a sub-confluent monolayer of M2-10B4 cells for 1 h at 37 °C. The supernatant was then removed, and cells grown in MEM + 2% NCS containing carboxy-methylcellulose for 4 days. Cells were fixed, stained with 0.5% methylene blue in 10% formaldehyde for 24 h and plaques in the monolayer counted.
Isolation of leukocytes and fibroblastic reticular cells
Mice were humanely killed and spleens and/or lymph nodes removed. FRCs from the spleen or lymph nodes were isolated by enzymatic digestion as described previously55. Spleen or lymph node leukocytes were isolated by mechanical disruption of tissues, except when CD44 expression on these cells was compared to that in stromal populations (for example, fibroblastic cells and endothelial cells), in which case the stromal isolation protocol55 was used. Prior to staining, red blood cells were lysed using an ammonium chloride–potassium lysis solution.
Cell staining and flow cytometric analysis
Cell surface staining of single-cell suspensions was performed using fluorescently conjugated antibodies in combination with pMHCI tetramers Ld-IE1168–176, or Db-M45985–993 for the identification of MCMV-specific CD8 T cells, Db-NP366–374 for the identification of IAV-specific CD8 T cells, or Kb-OVA257–264 for the identification of OVA-specific CD8 T cells. Dead cells were excluded using 4′,6-diamidino-2-phenylindole hydrate (DAPI) for live cells or FVS440UV (BD Biosciences) for fixed cells. For hyaluronic acid staining, cells were fixed in 2% paraformaldehyde (PFA) and stained overnight prior to analysis. Cells were acquired on a FACSymphony A3 cell analyser running FACSDiva (BD Biosciences), and data analysis performed with FlowJo software (BD Biosciences). Gating strategies are shown in Extended Data Fig. 10. Details of the antibodies used for flow cytometry are provided in Extended Data Table 3.
Immunofluorescence
IC-21 cells infected with MCMV (K181) were collected at 4 dpi and the cell suspension was pre-incubated on ice for 30 min with 10% normal goat serum (NGS) plus 2% FCS in PBS to block nonspecific reactivity. The cells were stained with anti-vCD44BP (M-627) followed by anti-mouse IgG biotin and Streptavidin-Alexa Fluor 488. After staining, cells were fixed in 4% PFA, dried onto glass slides and examined by epifluorescence microscopy (Olympus, BX60).
The FRC2 cell line or primary splenic FRCs were grown on glass coverslips or 96-well PhenoPlate (black, optically clear, flat bottom, tissue culture treated), infected with MCMV or ΔvCD44BP viruses for 24 h, fixed in 4% PFA and stained with antibodies, as indicated in figure legends. For vCD44BP immunofluorescent imaging, cells were stained with anti-vCD44BP (clone 7G5, 20 μg ml−1) for 1 h at 37 °C, fixed in 4% PFA for 15 min at room temperature, followed by incubation with biotin-conjugated anti-rat-IgG. Samples were then treated with a tyramide signal amplification kit (Invitrogen), followed by the addition of Alexa488-conjugated Streptavidin. Samples were permeabilized in 0.1% Triton X-100 before the addition of anti-CD44 antibodies, phalloidin and DAPI. Glass coverslips were mounted with Fluoromount Aqueous Mounting Medium (Sigma). Images were acquired using the following confocal instruments: Leica SP5 and SP8 at 1,024 × 1,024 pixels at 64× magnification with 8-bit sensitivity or Leica DMi8 Inverted Microscope with 8-bit sensitivity.
For immunofluorescence studies of spleen sections, mice were humanely killed, and spleens were excised and fixed overnight in periodate-lysine-paraformaldehyde that was prepared as described59. Spleens were then transferred into 30% sucrose for 24 h before embedding in Tissue-Tek OCT compound, frozen on isopentane over dry ice and stored at −80 °C. Cryostat sections (6–20 µm thick) were cut and air dried before fixation with −20 °C acetone and quenching with 50 mM ammonium chloride. Sections were blocked with 10% NGS and stained with primary antibodies overnight at 4 °C. The next day, sections were washed prior to staining with secondary antibodies for 1 h at room temperature. Images were acquired using a Leica DMi8 Inverted Microscope with 8-bit sensitivity, Nikon AX R Ti2-E confocal microscope, or Carl Zeiss LSM980 confocal microscope.
Image analysis was performed using the ImageJ software. Mean fluorescence intensities of CD11c, Xcr1, or 33D1 staining were quantified within splenic white pulp regions. The cell morphology index (perimeter2/4π area) was calculated as described8. Skeleton analysis was performed as described60. First, immunofluorescence images were cropped to include the podoplanin positive area of the white pulp. Images were then de-speckled, thresholded, converted into binary images, skeletonized and analysed using the Skeletonize (2D/3D) and Analyse Skeleton (2D/3D) plugins, respectively. Details of the antibodies used for immunofluorescence are provided in Extended Data Table 4.
3D migration assays
Primary FRCs were isolated from C57BL/6J or B6 Cd44−/− spleens and cultured as described55. Dendritic cells were expanded from bone marrow progenitors as described61, labelled with Cell Tracker Deep Red (Invitrogen) and seeded at a 5:1 ratio with FRCs into a matrix of 1.8 mg ml−1 Collagen I (Corning), 2.6 mg ml−1 Matrigel (Corning) and 10% FBS in alpha-MEM62. In some assays, FRCs or dendritic cells were pre-treated with vCD44BP–Fc (20 μg ml−1 vCD44BP–Fc for 40 min at 4 °C) and washed to remove unbound vCD44BP–Fc prior to co-culture. Cultures were imaged with the Opera Phenix Plus High-Content Screening System (PerkinElmer) for 4 h at 37 °C with 5% CO2. Migration of dendritic cells in contact with FRCs was tracked with ImageJ using the Manual Tracking plugin.
Quantification of CD44–vCD44BP co-localization
The FRC2 cell line was infected with MCMV or ∆vCD44BP for 24 h, stained with anti-CD44 and anti-vCD44BP (clone 7G5) antibodies, and cells analysed using an Amnis INSPIRE ImageStreamX instrument (Cytek Biosciences). Data analysis was performed using Image Data Exploration and Analysis Software (IDEAS) and rank-weighted co-localization was analysed using the co-localization pipeline in CellProfiler.
vCD44BP impact on adjuvant-induced LN expansion, immunization and influenza infection
Montanide adjuvant (Seppic) (25% diluted in PBS) was administered by subcutaneous injection in the neck scruff of mice. On days 1 and 3 post-adjuvant treatment, 15 µg of vCD44BP–Fc, control human TRAIL–Fc (a human protein that does not bind in the mouse), or PBS were administered by subcutaneous injection adjacent to the adjuvant injection site. At day 5 post-adjuvant treatment, lymph nodes that drain the injection site (cervical) were isolated, pooled and weighed prior to flow cytometric analysis. As a control, non-draining lymph nodes (inguinal) were also isolated and treated as above.
Antigen-specific immune responses were assessed by emulsifying 40 µg of OVA protein (Merck Life Science) with Montanideadjuvant. On days 1 and 3 post-adjuvant treatment, 15 µg of vCD44BP–Fc, hTRAIL–Fc or PBS were administered by subcutaneous injection adjacent to the adjuvant + OVA injection site. At day 6 post-adjuvant + OVA treatment, cervical and inguinal lymph nodes were isolated and analysed by flow cytometry.
On days −1, 0 and 3 relative to influenza infection, 50 µg of vCD44BP–Fc, control hTRAIL–Fc or PBS were administered by intravenous injection. At 10 dpi, the mediastinal lymph nodes were isolated and single-cell suspensions were prepared and analysed by flow cytometry.
Quantitative real-time PCR
Quantitative real-time PCR was performed using the SsoAdvanced Universal SYBR Green Supermix using a CFX Connect Real-Time System (Bio-Rad). The ribosomal protein L32 was used as the control housekeeping gene: forward, 5′-CATCGGTTATGGGAGCAAC-3′; reverse, 5′-GCACACAAGCCATCTACTCAT-3′. The following transcripts were detected: CCL19: forward, 5′-CCTGGGTGGATCGCATCA-3′; reverse, 5′-TGCCTTTGTTCTTGGCAGAA-3′, CCL21: forward, 5′-GCAAAGAGGGAGCTAGAAAACAGA-3′; reverse, 5′-TGGACGGAGGCCAGCAT-3′.
Statistical analysis
All data were analysed and graphed as mean ± s.e.m. or violin plots using Prism (GraphPad), unless otherwise stated. Statistical significance was determined using either a Mann–Whitney U-test or a Kruskal–Wallis test with a post hoc Dunn’s test for multiple comparisons, as detailed in the figure legends.
Software and algorithms
The software packages and algorithms used in this study are listed in Extended Data Table 5.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.