Animals
C57BL/6 mice and K18-hACE2 mice (strain: B6.Cg-Tg(K18-ACE2)2Prlmn/J) were purchased from the Jackson Laboratory. Fgaâ/â mice52 and Fggγ390â396A mice53 were obtained from J. Degen. Mice were housed under a 12âhâ12âh lightâdark cycle, 55â±â5% relative humidity at 20â±â2â°C with access to standard laboratory chow and water ad libitum. Both male and female mice were used. The mouse ages are indicated for each experimental procedure and were within 3 to 7 months of age. All infection experiments were performed at an AAALAC-accredited ABSL3 facility at Gladstone Institutes. All of the animal procedures were performed under the guidelines set by the Institutional Animal Care and Use Committee at the University of California, San Francisco.
Human plasma and PBMCs
Human citrated plasma (IPLASEATNAC50ML, 1151254) was purchased from Innovative Research. Fresh PBMCs (LP,FR,MNC,2B; 3118730 and 3112992) were purchased from AllCells. All human material used in the study is commercially available and no human participants were recruited.
SARS-CoV-2 recombinant trimeric spike protein production
The plasmid vector pCAGGS containing the SARS-CoV-2,WuhanâHuâ1 ectodomain spike gene with a deletion of the polybasic cleavage site (RRAR to A), two stabilizing mutations (K986P and V987P), a C-terminal thrombin cleavage site, T4 foldon trimerization domain and a hexahistidine tag (6ÃHis) was obtained from BEI Resources (deposited by F. Krammer)54. Recombinant spike was produced by transient transfection in CHO cells by Celltheon. Spike was purified by Ni2+-NTA affinity chromatography, eluted in phosphate-buffered saline (PBS) containing imidazole, buffer exchanged into 1Ã PBS and purified by size-exclusion chromatography (Superdex 200 column).
Plasma clot formation assay
Fibrin polymerization in a plasma clot assay was measured by turbidity17. In brief, healthy donor citrated human plasma (Innovative Research) was diluted 1:3 in 20âmM HEPES. Recombinant spike was buffer-exchanged into 20âmM HEPES, pHâ7.4, 137âmM NaCl (Amicon concentrators, 100âkDa cut-off). Equal volumes (50âµl) of plasma and buffer-exchanged spike were incubated at 25â°C for 15âmin. Clotting was initiated by 0.25âUâmlâ1 thrombin (Sigma-Aldrich) and 20âmM CaCl2. The final concentrations were 1:12 plasma, 0.75âμM spike, 0.25âUâmlâ1 thrombin, 20âmM CaCl2. Turbidity was measured at 340ânm every 15âs for 30âmin on the SpectraMax M5 microplate reader (Molecular Devices) using SoftMax Pro v.5.2 (Phoenix Technologies).
SEM analysis of fibrin clots
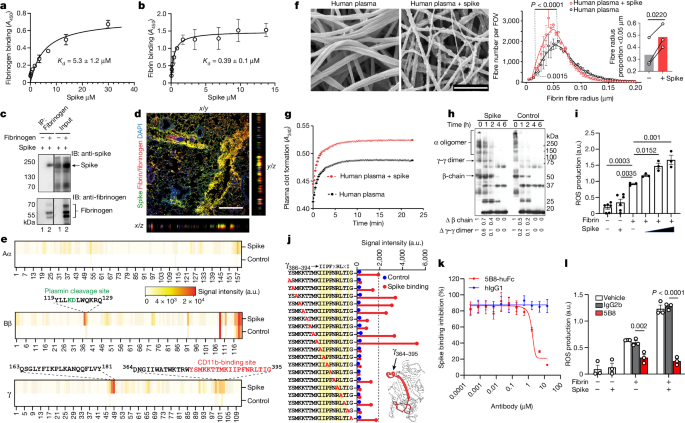
Healthy donor citrated human plasma was diluted 1:3 in 20âmM HEPES buffer, pHâ7.4; 15âμl of diluted plasma was mixed with 15âμl of recombinant spike that was buffer-exchanged into 20âmM HEPES and 137âmM NaCl (Amicon concentrators, 100âkDa cut-off) using a low concentration of NaCl to maintain spike solubility and stability. Then, 25âμl of this mixture was pipetted onto 5âmmâÃâ5âmm silicon wafers (Ted Pella) and incubated for 15âmin at 37â°C in a humidified tissue culture incubator. Then, 25âµl of a CaCl2 and thrombin solution in 20âmM HEPES was added in the centre of the wafer and allowed to polymerize at 25â°C for 2âh. The final concentrations were as follows: plasma 1:12, 0.9âμM spike, 0.25âUâmlâ1 thrombin, 20âmM CaCl2. Buffer was used instead of spike for vehicle control. Clots on wafers were placed onto ice, washed twice for 10âmin each with ice-cold EM-grade 0.1âM cacodylate buffer, pHâ7.4, and fixed with cold EM-grade 2% glutaraldehyde (Electron Microscopy Sciences). The samples were rinsed three times for 5âmin in Millipore-filtered, double-distilled water; dehydrated in an ethanol series (20%, 50%, 70%, 90%, 100%, 100% for 2âmin each); and critical-point dried with CO2. The samples were sputter coated with a thin layer of goldâpalladium and imaged on the Zeiss Merlin field-emission SEM at 3.0âkeV and a secondary electron detector.
Images at a magnification of Ã4,000 were captured across the sample, then were converted to 8-bit using NIH ImageJ (v.1.50). After pixel to μm scaling, each image was cropped into two or three FOVs (8âÃâ8âμm) using NIH DiameterJ as described previously55. The surface plot plug-in in ImageJ generated topographical maps of SEM images. In brief, the best segmentation algorithm was pre-selected based on side-by-side comparison of images before quantification. The Mixed Segmentation (M1 through M3) built in DiameterJ Segment provided the most accurate representation of the fibres to be quantified. The same segmentation method and variant was used across all test conditions and images. Each segmented image was manually edited using ImageJ to ensure complete representation of segmented fibres. The edited images were batch processed using DiameterJ 1-108 (orientation analysis not selected). Fibre radius and intersection densities were collated from each batch. Data from 8â10 FOVs per sample were used for group analysis. Fibre radius distribution in Fig. 1f was calculated using FOVs from all images collected to assess the distribution across the dataset. Fibre radius proportion was statistically analysed based on three biologically independent experiments in Fig. 1f and the quantification and statistical analysis of the individual images from these experiments is shown in Extended Data Fig. 3c. Samples with collapsed fibres due to potential SEM critical-point drying technical artifacts were excluded from further analysis.
For quantification of the fibrin clots by SEM, at each radius, the difference in log-transformed odds ratio of detecting fibres (among all the views in a given image) with the chosen radius under spike versus control conditions was estimated across all images. The log-transformed odds ratio at each radius was estimated using generalized linear mixed-effects models, with the family argument set to binomial and implemented in glmer function in the lme4 (v.1.1-27) package in R56, in which the image source for the observations is modelled as a random effect. The P values were corrected for multiple testing using the Holm procedure57. In Fig. 1f, the P value represents the significance at each radius across the range of the radii between the two vertical dotted lines. The solid lines represent the best loess fit curves with span parameter set to 0.45.
Fibrinogen- and fibrin-coated ELISA plates
Fibrinogen- and fibrin-coated plates were prepared as described previously17. In brief, human plasminogen-free fibrinogen (EMD Millipore) was further diluted to 25âµgâmlâ1 by adding 20âmM HEPES buffer, pHâ7.4 for coating fibrinogen plates or 20âmM HEPES buffer pHâ7.4 with 1âUâmlâ1 thrombin (Sigma-Aldrich) and 7âmM CaCl2 for fibrin-coated plates. Coating was performed for 1.5âh at 37â°C using 96-well MaxiSorp plates (Thermo Fisher Scientific) and fibrin-coated plates were dried at 37â°C overnight as described previously17.
Recombinant SARS-CoV-2 spike protein binding on fibrin or fibrinogen
Fibrin- or fibrinogen-coated 96-well plates were washed with wash buffer (0.05% Tween-20 in PBS), and incubated with blocking buffer consisting of wash buffer with 5%âbovine serum albumin (BSA) (Omnipure, Thermo Fisher Scientific) for 1âh at 25â°C. Serial dilutions of recombinant spike or S1(N501Y) were made in binding buffer (wash buffer containing 0.5% BSA). Recombinant spike or S1(N501Y) was added to the wells and incubated for 2âh at 37â°C. After washing five times with wash buffer, rabbit polyclonal anti-6à His tag antibody (ab137839, Abcam, 1:1,000) was added to the plates and incubated for 1âh at 25â°C. After washing, goat anti-rabbit IgG H&L (conjugated with horse radish peroxidase, HRP) (ab205718, Abcam, 1:1000) in wash buffer was added for 1âh at 25â°C. After the final wash, the HRP substrate 3,3â²,5,5â²-tetramethybenzidine (TMB; Sigma-Aldrich) was added into the wells. The reaction was quenched by adding 1âN hydrochloric acid, and the absorption was measured at 450ânm. Nonlinear regression curves were analysed using Graph Pad Prism 9 software to calculate Kd values using a one-site binding model.
Fibrinogen peptide array and spike-binding epitope mapping
A custom PepStar Multiwell Fibrinogen Peptide Array comprising a synthetic peptide library with 390 15-mer peptides representing overlapping peptide scans (15/11) of the α, β and γ fibrinogen chains (UniProt: FIBA, P02671; FIBB, P02675; FIBG, P02679) was generated by JPT Peptide Technologies. The arrays were hybridized with recombinant-His-tagged trimeric spike (1âµgâmlâ1 in blocking buffer) for 1âh at 30â°C. The His-tag peptide (AGHHHHHH) was also immobilized on the peptide microarray as an assay control. Microarray slides were incubated for 1âh at 30â°C with Alexa 647 anti-6ÃHis monoclonal antibody (MA1-135-A647, Invitrogen) diluted to 1âµgâmlâ1 in blocking buffer and dried. Before each step, microarrays were washed with washing buffer, 50âmM TBS-buffer including 0.1% Tween-20, pHâ7.2. The assay buffer was LowCross buffer (Candor Bioscience). The slides were washed, dried and scanned with a high-resolution laser scanner at 635ânm to obtain fluorescence intensity profiles. The images were quantified to yield a mean pixel value for each peptide. To assess non-specific binding to the peptides and assay performance, a control incubation with secondary antibody was performed in parallel on each slide. The resulting images were analysed and quantified using spot-recognition software (GenePix, Molecular Devices). For each spot, the mean signal intensity was extracted (between 0 and 65,535 arbitrary units). Heat maps were computed and fluorescence intensities were colour-coded. Binding peptides were mapped onto the fibrinogen crystal structure (Protein Data Bank (PDB): 3GHG) using UCSF Chimera58. For the spike peptide array, 1, 0.1 or 0.01âµgâmlâ1 His-tagged recombinant human fibrinogen γ chain (Novus Bio) was hybridized with the SARS-CoV-2 spike Glycoprotein Variant Collection Peptide Microarray (JPT). Binding was detected using an anti-His secondary antibody conjugated to Alexa 647. Non-specific binding was detected using an anti-His secondary antibody only. Separately, 1, 0.1 or 0.01âµgâmlâ1 Alexa-647 fibrinogen (Invitrogen) was hybridized onto the spike Glycoprotein Variant Collection Microarray, and peptide binding was directly detected by fluorescence intensity in relative light units (RLU). A heat map was generated by using raw RLU for side-by-side comparison. Spike glycoprotein binding sites on fibrinogen were mapped using the PDB (6VSB).
Peptide alanine scanning
Alanine scanning was performed with custom PepStar Multiwell microarrays (JPT) containing 60 peptides representing Ala substitutions of each residue on fibrinogen peptide γ377â395 (YSMKKTTMKIIPFNRLTIG). Human full-length IgG and His-tagged peptides were co-immobilized on the microarray slides as controls. His-tagged spike was applied at five concentrations (from 10âμgâmlâ1 to 0.001âμgâmlâ1) and incubated for 1âh at 30â°C. Two fluorescently labelled secondary antibodies specific to the His tag were added separately for 1âh. Washing and detection was performed as described above and data were analysed with respect to the original peptide. The signal after Ala substitution indicated whether a residue was involved in binding to spike.
Structure preparation and homology modelling
The crystal structure of human fibrinogen (PDB: 3GHG) was fixed using the Structure Preparation application of the Compute module of MOE. The crystal structure of SARS-CoV-2 spike (PDB: 6VSB) has missing structural information for flexible loops. To correct these, the Homology Model application in the Protein menu of MOE 2022.02 software (Chemical Computing Group) was used, which includes: (1) initial partial geometry specification; (2) insertions and deletions; (3) loop selection and sidechain packing; and (4) final model selection and refinement. Homology models were inspected using MOEâs Protein Geometry stereochemical quality evaluation tools. The spike crystal structure (PDB: 6VSB) was prepared by assigning protonation and ionization states.
Docking and calculation of energies of docked complexes
Docking of two proteins was performed by Dock application of Compute module of MOE, using the Protein-Protein function. The application generates a collection of docked configurations from the pool of possible binding positions using the rigid-body docking. To complete a docking procedure, the binding sites were identified based on the peptide array described above. Three potential binding sites were chosen for fibrinogen: (1) 119YLLKDLWQKRQ129 in the β-chain; and, in the γ-chain, (2) 163QSGLYFIKPLKANQQFLVY181 and (3) 364DNGIIWATWKTRWYSMKKTTMKIIPFNRLTIG395. For the ligand (spike protein) five sites were selected. NTD binding region: (1) 37YYPDKVFRSSVLHSTQDLFLPFFSNVTWFHAIHVSGTNGTKRFDNPVLPFNDGVYFASTEKSNIIRG103, (2) 229LPIGINITRFQTLLALHRSYLTP251 and (3) 305SFTVEKGIYQTSNF319; RBD region: (4) 341VFNATRFASVYAWNR355; and S2 domain: (5) 1049LMSFPQSAPHGVVFL1063. After receptor, ligand and docking sites were defined, parameters of Dock application of the Compute module of MOE were set to: refinement –Rigid Body, Poses –10. The application created 10 poses, analysed output scores, ligand docking energies and docked poses, and detected the best one; intermediate poses also are saved in a docking database file.
During the docking calculations the program presents 10 best energy complexes. After that, each of the complexes undergone the additional calculations of energy. A computational alanine scan of the fibrinogen molecules in each complex was also conducted with each of the residues in fibrinogen that were experimentally substituted to alanine were computationally substituted to alanine and modelled. The best model was selected on the basis of the lowest docking energy. The computational alanine scan generated the values of correlations between all values of energy for each amino acid substitution and experimental values of the parameter used for estimating the influence of each amino acid. The residues involved in the interaction of this computationally predicted complex were analysed using LigPlot+ v.2.2.
i.v. injection of labelled spike S1(N501Y) and fibrinogen
Spike S1(N501Y) (AcroBiosystems) (20âμg) dissolved in 0.1âM PBS was fluorescently labelled using the Alexa Fluor 647 conjugation kit lightning link (Abcam). The Alexa-Fluor-647-labelled spike S1(N501Y) had a concentration of 1âmgâmlâ1. Retro-orbital injections of 0.1âml of PBS solution containing 20âμg Alexa-647-conjugated spike S1(N501Y) and 30âμg Alexa-546-labelled human fibrinogen (Invitrogen) were performed under isoflurane anaesthesia (1âml insulin syringe with a 30-gauge needle). The mice were perfused at 1âday after injection with heparinized PBS and fixed with 4% paraformaldehyde (PFA) and lungs were collected for clearing.
3DISCO clearing and light-sheet imaging
3DISCO lung tissue clearing was performed as described previously59. Mouse lungs were placed into a 20âml scintillation glass vial and incubated in 20âml of THF (Tetrahydrofuran, Roth, CP82.1) gradient in distilled water in a fume hood with gentle shaking at 50% once, 70% once, 80% once and 100% twice for 6âh for each step, followed by 3âh in dichloromethane (DCM, Sigma-Aldrich, 270997). The samples were immersed in BABB solution (benzyl alcohol + benzyl benzoate 1:2 (v/v), Sigma-Aldrich, 24122 and W213802) until optical transparency. Lung tissues were imaged using Imspector Pro v.7.0.98 and the LaVision BioTec Ultramicroscope II light-sheet microscope in a quartz cuvette filled with ethyl cinnamate (ECi) (Sigma-Aldrich). For imaging, MVX10 zoom body (Olympus) with a Ã2 objective (pixel size, 3.25âµm for x and y) at magnification from Ã0.63 up to Ã1.6 was used. Up to 1,400 images were taken for each lung using a z-step size of 3.5âµm, and light-sheet numerical aperture of 0.111 NA. Band-pass emission filters (mean nm/spread) were used, depending on the excited fluorophores: 525/50 for autofluorescence; 595/40 for AF546; and 680/30 for AF647. The exposure time was 10âms for single channel and 25âms for multichannel acquisition. Imaris v.9.5.0 (Bitplane) was used for 3D rendering. Pixel dimensions were updated from the non-reduced 16-bit image metadata. Surface objects in Imaris was used to 3D render focal depositions and spike distribution in representative volumetric ROIs.
Plasmin digestion of fibrin
Before clotting, 3âμM fibrinogen was incubated with 9âμM recombinant spike protein at 37â°C for 1âh in 20âmM HEPES, pHâ7.4, 137âmM NaCl, 5âmM CaCl2. Thrombin was added at a final concentration of 1.5âUâmlâ1. Fibrin clots were allowed to form in Eppendorf tubes for 2âh at 37â°C. Then, 5âμl of 100âμgâmlâ1 plasmin (Millipore) was added to each tube on top of the clot. All of the samples were incubated at 37â°C for 0, 1, 2, 4 and 6âh; digestion was quenched by adding sodium dodecyl sulfateâpolyacrylamide gel electrophoresis (SDSâPAGE) loading buffer with reducing agent. The samples were heated at 85â°C for 20âmin, and aliquots (equivalent to 100âng fibrinogen) were separated by SDSâPAGE on 4â12% Bis-Tris gels, transferred to PVDF membranes and analysed for anti-human fibrinogen (F4200-06, US Biological, 1:2,000) by western blotting. The band intensities of each protein species (that is, γâγ dimer, β-chain) were analysed using ImageJ and normalized to the corresponding bands at the 0âh timepoint. The loading control for the western blot is the timepoint 0 before the addition of plasmin to the fibrin clot.
Competitive ELISA of 5B8 versus the spike for binding to fibrin
A 5B8-huFc antibody was synthesized by Fc swap of the mouse IgG2b Fc of 5B818 with human IgG1 Fc. 96-well ELISA plates (Greiner) were coated with 25âμgâmlâ1 IgG-depleted fibrin and incubated in blocking buffer as indicated for binding assays for 1âh before addition of 50âμl per well of 5B8-huFc antibody. Human plasminogen-free fibrinogen was depleted from IgG as described previously17. The antibody was diluted at threefold concentrations from 0.0002âμM to 15âμM in PBS with 0.5% BSA and 0.05% Tween-20 (diluent). For the competition ELISA without preincubation, 5B8-huFc was incubated together with 150ânM trimeric spike in diluent (100âμl total volume) for 2âh at 37â°C on fibrin plates. For the ELISA with antibody preincubation, 50âμl of 5B8-huFc was incubated on fibrin plates for 2âh at 37â°C, followed by addition of 50âμl of 150ânM trimeric spike to the antibody and incubation for 2âh at 37â°C. This was followed by incubation with HRP-coupled anti-His tag antibody (MAB050H, R&D Systems, 1:2,000) for 1âh at 25â°C. The ELISA was developed by incubation with TMB/E substrate (Chemicon-Millipore), and the absorbance was measured at 450ânm using the Synergy H4 plate reader (BioTek).
ROS detection
BMDM culture and ROS detection using 5âµM DHE (Invitrogen) were performed as described previously17,60. In brief, cells were plated on 96-well black μ-clear-bottomed microtitre plates (Greiner Bio-One) precoated with 12.5âµgâmlâ1 fibrin with or without recombinant spike (0.168, 1.68 and 3.36âµM), spike PVs or bald PVs. For fibrin inhibition, 5B8 or IgG2b (each 20âμgâmlâ1) (MPC-11, BioXCell) was added in fibrin with or without 3.36âµM recombinant spike-coated wells 2âh before plating. Cells were incubated on fibrin for 24âh and DHE fluorescence was detected at 518ânm/605ânm using the SpectraMax M5 microplate reader. As macrophage activation can be influenced by cell culture conditions, heat-inactivated fetal bovine serum and macrophage colony-stimulating factor were batch tested as described previously60. As the activity of PVs can be influenced by freezeâthaw cycles, all of the experiments were performed with virions that had been freshly thawed and kept at 37â°C. Refrozen virion samples were not used.
Immunoprecipitation
To test interaction of fibrinogen with His-tagged spike, the Pierce co-immunoprecipitation kit (Thermo Fisher Scientific) protocol was used with original immunoprecipitation/lysis buffer and modifications. Spike and fibrinogen were mixed at a molar ratio of 2:1 in 800âμl of immunoprecipitation buffer (50âmM Tris, pH 8.0, 5% glycerol, 1% NP-40, 100âmM NaCl) supplemented with 100 à EDTA-free Halt protease inhibitor (Thermo Fisher Scientific) and then rotated at 37â°C for 1âh. Resin beads conjugated with the anti-fibrinogen antibody (SAFG-AP, Enzyme Research Laboratories, 1:1,000) were added to the mixture and rotated at 37â°C for another 2âh. The bound proteins were eluted in 60âµl of EB solution and neutralized with 1/10 volume of 1âM Tris, pHâ9.0. The wash buffer and EB solution were warmed to 37â°C in advance. The eluted proteins were separated by SDSâPAGE on 4â12% gels, transferred to PVDF membranes (Invitrogen) and incubated with rabbit anti-spike antibody (632604, GeneTex, 1:1,000) and sheep anti-fibrinogen antibody (SAFG-AP, Enzyme Research Laboratories, 1:1,000) and then with HRP-conjugated anti-rabbit (111-035-144, Jackson ImmunoResearch; 1:10000) and anti-sheep (HAF016, R&D Systems; 1:5,000) secondary antibodies. For immunoprecipitation of spike PVs, spike antibodies (GTX635693, GeneTex; 1:1,600) recognizing SARS-CoV-2 spike (S2) were used. For spike PV immunoblot, anti-spike (632604, GeneTex, 1:1,000) and anti-p24 Gag (detecting p55, 1:100) antibodies donated to the Greene laboratory by Beckman Coulter61 and anti-Vpr (8D1, Cosmo Bio, 1:200) antibodies were used. Protein bands were detected using Immobilon Forte Western HRP substrate (Sigma-Aldrich) and the ChemiDoc imaging system (Bio-Rad).
SARS-CoV-2 culture and in vivo infection
To assess SARS-CoV-2 infection in vivo, viral stocks of SARS-CoV-2 B.1.351 (Beta) and SARS CoV-2 B.1.617.2 (Delta) were prepared on Vero cells expressing transmembrane protease serine 2 (TMPRSS2) and ACE2 (Vero-TMPRSS2-ACE2)47 provided by A. Creanga and B. Graham at NIH and stored at â80â°C until used. Experiments involving Beta were performed on female and male WT C57BL/6, Fgaâ/â and Fggγ390â396A mice (6â7 months of age). The Beta strain contains the K417Y, E484K and N501Y substitutions in the spike RBD and binds to mouse ACE2 inducing active infection in a range of experimental mouse strains62,63,64. Experiments using Delta were performed on female and male 4â5 month old K18-hACE2 mice. For the infection, the animals were anaesthetized using 100âmg per kg ketamine mixed with 10âmg per kg xylazine through intraperitoneal injection. Anaesthetized mice received i.n. administration of an infectious inoculum of virus in 50âμl of serum-free DMEM. For each experiment, lung and brain tissues were collected. Left lung lobes and one brain hemisphere from each animal were placed in 4% PFA for fixation and histological processing. The remaining lung tissue was roughly chopped and processed for homogenates in prefilled zirconium bead tubes (Benchmark Scientific). Homogenates were stored at â80â°C. The remaining brain hemispheres were flash-frozen and stored at â80â°C. All aspects of this study were approved by the office of Environmental Health and Safety at UCSF before initiation. Work with SARS-CoV-2 was performed in a biosafety level 3 laboratory by personnel equipped with powered air-purifying respirators.
Plaque assay
Lung homogenates were assessed for viral concentration by plaque assay. In brief, Vero-TMPRESS2-ACE2 cells were plated onto 12-well plates at a concentration of 2âÃâ105 cells per well. Homogenates were added to the cells in a dilution series of 101, 102, 103, 104, 105 and 106 in serum-free DMEM. The homogenate dilutions were incubated on the cells for 1âh, and the media in the wells was then overlaid with 2.5% Avicel (Dupont, RC-591). Cells were incubated for 72âh, then the overlay was removed and the cells were fixed in 10% formalin for 1âh, and stained with crystal violet to visualize PFU.
Production of spike PVs
HEK293T cells (3.75âÃâ106) were plated in a T175 flask and transfected 24âh later with 90âμg of polyethyleneimine (PEI; Sigma-Aldrich), 30âμg of HIV-1 NL4-3 â Env eGFP (NIH AIDS Reagent Program) or 3.5âμg of pCAGGS SARS-CoV-2 trimeric spike glycoprotein (NR52310, BEI Resources) in a total of 10âml of Opti-MEM medium (Invitrogen). The next day, the medium was replaced with DMEM10 complete medium, and the cells were incubated at 37â°C in 5% CO2 for 48âh. The supernatant was then collected, filtered with 0.22âµm Steriflip filters (EMD, Millipore) and ultracentrifuged at 25,000ârpm for 1.5âh at 4â°C. The concentrated supernatant was removed, the pellets (viral particles) were resuspended in cold 1à PBS containing 1% fetal bovine serum and aliquots were stored at â80â°C in a biosafety level 3 laboratory. For the production of control viral particles not expressing the spike glycoprotein (bald), the same procedure was used but with the omission of the pCAGGS SARS-CoV-2 spike vector transfection. HIV Env pseudotyped viral particles were also produced with the same procedure, using an HIV89.6 Env dual tropic (X4 and R5) expression vector (NIH AIDS Reagent Program) instead of the spike expression vector.
In vivo administration of SARS-CoV-2 spike PVs
Mice were anaesthetized with isoflurane and spike PVs or bald PVs (control) (100âµl) were slowly injected into the retro-orbital plexus with a BD 0.3âml insulin syringe attached to a 29-gauge needle. After 3âmin, the needle was slowly withdrawn, and the mice were allowed to recover. As the activity of PVs can be influenced by freezeâthaw cycles, all of the experiments were performed with virions that had been freshly thawed and kept at 37â°C. Refrozen virion samples were not used. SARS-CoV-2 spike PVs were administered to 3- to 4-month-old mice.
5B8 penetration in the CNS and target engagement after SARS-CoV-2 infection
C57BL/6 mice (4â5 months of age) were infected with 104 PFU of SARS-CoV-2 B.1.351 (Beta). On 5 and 7 d.p.i, mice were given intraperitoneally 30âmg per kg of the 5B8-huFc antibody. On 7 d.p.i, mice were perfused with saline followed by fixation with 4% PFA. Subsequently, the brains were post-fixed in the same fixative and cryoprotected in 30% sucrose. The brain hemispheres were frozen in OCT and sectioned (10âµm sections). Sagittal brain sections were incubated with 0.1% Sudan Black (dissolved in 70% ethanol) for 10âmin, permeabilized/blocked with 3% BSA and 3% NDS in PBS containing 0.1% Triton X-100 for 1âh. The sections were incubated overnight with an antibody to fibrinogen (1:2,000), followed by Alexa Fluor 594 donkey anti-rabbit IgG (1:1,000; Jackson ImmunoResearch) for 1âh. To detect 5B8-huFc antibody in the brain, the sections were stained with F(abâ²)2-donkey anti-human IgG (H+L) cross-adsorbed secondary antibody, FITC (ab102424, Abcam, 1:300) for 1âh. The sections were covered with glass coverslips, sealed with ProLong Diamond Antifade Mounting reagent (Thermo Fisher Scientific) and kept at 4â°C until imaging.
Fibrin 5B8 antibody treatment
For prophylactic pharmacological treatment of SARS-CoV-2 B.1.351 (Beta) infection, anti-fibrin antibody 5B817 or an isotype-matched IgG2b (MPC-11, BioXCell) control were administered intravenously by retro-orbital injection at 30âmg per kg in 5- to 6-month-old C57BL/6 mice. Then, 1âh later, the mice were given 104 PFU of Beta through the i.n. route in a final volume of 50âμl. Beta-infected mice were euthanized at 3âdays for histological analysis. For SARS CoV-2 B.1.617.2 (Delta) infection, 4- to 5-month-old K18-hACE2 mice were given 5B8 or IgG2b intravenously through retro-orbital injection at 30âmg per kg 1âh before Delta infection and every 48âh intraperitoneally, and were euthanized at 3 d.p.i. For therapeutic treatments, 5B8 or IgG2b were given intraperitoneally at a dose of 30âmg per kg at 1âd.p.i. with 103 PFU of Beta in 5- to 6-month-old C57BL/6 mice or Delta in 4- to 5-month-old K18-hACE2 mice as described above, and every 48âh thereafter, intraperitoneally. The animals were euthanized at 7 or 9 d.p.i. For spike PVs, 5B8 or IgG2b isotype control were given intravenously to C57BL/6 mice by retro-orbital injection at 30âmg per kg 15âmin before injection of PVs. Generation of 5B8 and dose of administration have been described previously17. Administration of mouse monoclonal antibodies intraperitoneally provides sustained release of antibody into the bloodstream and thus is commonly used to assess preclinical efficacy for antibodies that will eventually be delivered intravenously in the clinic65,66,67.
Histology and immunohistochemistry
Histopathological analysis in mouse lung and brain was performed on frozen or paraffin sections17,68,69. Serial sections were not collected in the study. Lung sections were stained with haematoxylin and eosin and trichrome. The following antibodies were used: rabbit anti-SARS-CoV-2 nucleocapsid (GTX135357, GeneTex, 1:500), mouse anti-SARS-CoV-2 spike (1A9, GeneTex, 1:100), sheep anti-fibrinogen (F4200-06, US Biological, 1:300), rabbit polyclonal anti-fibrinogen (gift from J. Degen, 1:500), rat anti-mouse/human Mac2 (M3/38, Cedarlane, 1:500), mouse anti- gp91phox (53/gp91-phox, BD Biosciences, 1:500), rat anti-mouse CD335 (NKp46) (29A1.4, BD Biosciences, 1:500), mouse anti-NK1.1 (PK136, Invitrogen, 1:250) and rabbit anti-granzyme A (PA5-119160, Invitrogen, 1:500). Brains were cut with a cryostat into 30-μm-thick frozen sections for free-floating immunostaining. The following antibodies were used: rabbit anti-IBA1 (019-19741, Wako, 1:1,000), rat anti-mouse CD68 (FA-11, BioLegend, 1:500), guinea pig anti-NeuN (A60, Sigma-Aldrich, 1:500), rat anti-myelin basic protein (ab7349, Abcam, 1:100) and rabbit anti-calbindin (CB38a, Swant; 1:5,000). The tissue sections were washed in PBS and incubated in a blocking and permeabilization buffer consisting of PBS supplemented with 0.2% Triton X-100 and 5% BSA for 1âh at 25â°C. For mouse primary antibodies, the sections were incubated in M.O.M. (Mouse on Mouse Immunodetection Kits, Vector Laboratories) mouse IgG blocking reagent diluted in PBS containing 0.2% Triton X-100 and 5% BSA, and then with M.O.M. diluent for 5âmin at room temperature. The sections were rinsed twice with PBS containing 0.1% Triton X-100 and incubated overnight with primary antibodies at 4â°C. All of the tissue sections were washed with PBS containing 0.1% Triton X-100 and incubated with the following secondary antibodies: goat anti-rabbit Alexa Fluor 488 (A-11008, Thermo Fisher Scientific, 1:1,000), goat anti-mouse Alexa Fluor 568 (A-110041, Thermo Fisher Scientific, 1:1,000) or goat anti-rat Alexa Fluor 647 (A-21247, Thermo Fisher Scientific, 1:1,000), and stained with DAPI. The sections were mounted on frosted microscopy slides (Thermo Fisher Scientific), covered with glass coverslips, sealed with ProLong Diamond Antifade Mounting reagent (Thermo Fisher Scientific) and kept at 4â°C until imaging.
Confocal microscopy
Tissue sections were imaged using a laser-scanning confocal microscope FLUOVIEW FV3000RS âSnow Leopardâ (Olympus) or Fluoview FV1000 (Olympus), a 40 Ã and 0.8 NA water-immersion lens or 60Ã oil-immersion UPLSAPO objective (NAâ=â1.35) and FV31S-SW software v.2.3.2.169 (Olympus). Individual channels were captured sequentially with a 405ânm laser and a 430/70 spectral detector for DAPI, a 488ânm laser and a 500/40 spectral detector for Alexa Fluor 488, a 561ânm laser and a 570/620 high-sensitivity detector for Alexa Fluor 568, and a 650-nm laser and a 650/750 high-sensitivity detector (Olympus TruSpectral detector technology) for Alexa Fluor 647. Captured images were processed with Fiji v.2.1.0/ImageJ v.1.53c.
Image analysis
To analyse microglia after stereotaxic injections of fibrinogen, spike or PVs, the corpus callosum within five rostrocaudally spaced coronal brain sections was selected for quantification17. To quantify IBA1, CD68, calbindin or NeuN+ cells in mice infected with Beta or Delta, three areas in the hippocampus (for IBA1 or CD68) or two areas in the cortex (for calbindin or NeuN) were selected on three mediolaterally spaced sagittal brain sections, ensuring consistency in anatomical regions per mouse. For lung pathology in Beta-infected mice, six or seven representative areas were chosen from three lung sections. N protein-positive areas were selected for collagen quantification. Lung pathology in mice injected with PVs was performed on five representative areas selected from three lung sections. Immunostained cells were counted with Jupyter Notebook in Python 3. In brief, an arbitrary threshold was manually set and used for all images in the dataset. The total number of cells per image was estimated using the function peak_local_max from the open-source skimage Python image-processing library, which returns the coordinates and number of local peaks in an image (https://scikit-image.org/docs/dev/api/skimage.feature.html#skimage.feature.peak_local_max). Fibrinogen immunoreactivity was quantified using Fiji (ImageJ) as described previously70. Python image processing was used to colocalize fibrinogen and spike protein in lung tissues. In brief, a Jupyter Notebook was written to estimate the amount of fluorescence signal overlap between spike and fibrinogen in lung tissues. The Ostu filter from the skimage Python image-processing library was used to threshold each image labelled with spike and fibrinogen (https://scikit-image.org/docs/0.13.x/api/skimage.filters.html#skimage.filters.threshold_otsu). After thresholding, each set of images was compared, and pixels were compartmentalized into 4 categories: spike and fibrinogen overlap, spike signal only, fibrinogen signal only and no signal. In each image, the total number of pixels in an image and the number of pixels with signal for spike only, fibrinogen only or both were computed. Correlations were calculated using FOVs from all images collected as indicated in Extended Data Figs. 1b,c and 9f to assess the distribution across the dataset. All images selected for the figures are representative of the quantification of immunostaining for each experimental group.
Bulk RNA-seq
Lungs (3âd.p.i.) were isolated and snap-frozen with liquid nitrogen and stored at â80â°C. RNA samples were isolated using the RNeasy Plus Mini Kit (Qiagen). Generation of cDNA, sequencing, quality control of raw count, mapping and counting was performed as described21,60. The samples used for gene expression analysis were confirmed for viral load by quantitative PCR in lung tissue for expression of N5 specific for Beta variant. Samples with poor RNA quality or no viral load were excluded from further analysis. All of the samples that passed RNA quality control were included in the study. A minimum of three replicates per group was used, and genes with less than 0.1 counts per million (CPM) were filtered out from the study. Normalization was then performed using calcNormFactors, and differentially expressed genes were determined using edgeR71. The false-discovery rate (FDR) was calculated using the BenjaminiâHochberg method. For NK cell RNA-seq, adjusted Pâ<â0.1 (two-sided quasi-likelihood F-test with BenjaminiâHochberg correction) was used for visualization in Fig. 3a. The CPM of each gene was normalized across all of the samples to generate z-scores for heat maps of gene expression. Differentially expressed genes significantly changed in uninfected mice were not included in the analysis. For pathway analysis, gene lists were ranked using log2-transformed fold change of differentially expressed gene between two groups. Fibrin-induced macrophage scRNA-seq data were obtained from ref. 21 (GSE229376). GSEA was performed using GSEA v.4.2.3 with 1,000 times permutation and collapsing mouse genes to the chip platform Mouse_Gene_Symbol_Remapping_Human_Orthologs_MSigDB.v7.5.1.chip. The MSigDB gene sets: H: Hallmark and C2: CP: Canonical pathways (KEGG, REACTOME, WikiPathways) were used for pathway analysis. The fibrin NK suppression network was generated using Cytoscape (v.3.7.2)72. Using differentially altered pathways generated by GSEA (described earlier), the network was visualized using the default setting of EnrichmentMap.
NK cell depletion and characterization
NK cells were purified from splenocytes of C57BL/6 mice using the NK cell isolation kit (Miltenyi Biotec). NK cells were stimulated with IL-15 (50ângâmlâ1, BioLegend) for 4âdays with or without fibrin. Flow cytometry staining and analyses were performed as described previously21,60. For NK cell surface and intracellular staining, NK cell suspensions were first incubated with TruStain FcX PLUS (S17011E, BioLegend) for 15âmin at 4â°C, then stained with surface markers for 30âmin at 4â°C. Cells were then fixed and permeabilized using the BD Fixation/Permeabilization Kit (554714, BD). Intracellular markers were incubated for 1âh at 4â°C and analysed using the LSR Fortessa flow cytometer (BD Biosciences) the same day. For IFNγ staining, NK cells were incubated with phorbol 12-myristate 13-acetate (P8139, Sigma-Aldrich) and ionomycin (I0634, Sigma-Aldrich) for 4âh in the presence of brefeldin A (B7651, Sigma-Aldrich) followed by surface staining and fixation/permeabilization protocol described above. Anti-IFNγ antibodies were incubated in perm/wash buffer overnight, and then analysed with LSR Fortessa flow cytometer (BD Biosciences) the same day. Antibodies were as follows: NK1.1-FITC (S17016D, BioLegend, 1:200), IFNγ-PE (XMG1.2, BioLegend, 1:200), granzyme B-PerCP/Cy5.5 (QA16A02, BioLegend, 1:200), Ki-67-PE (16A8, BioLegend,1:200), CD45-Brilliant Violet BUV737(30-F11, BD, 1:200), CD11b-Brilliant Ultraviolet 395 (M1/70, BD, 1:200), CD335-Brilliant Violet 421 (clone 29A1.4, BioLegend,1:100), CD54-PE (YN1/1.7.4, BioLegend, 1:200), CD314-APC (CX5, BioLegend, 1:200), LIVE/DEAD Fixable Aqua Dead Cell Stain Kit (L34957, Thermo Fisher Scientific, 1:500). All data were processed using FlowJo v.10.7.1 (BD Biosciences). Doublets and dead cells were excluded before analysis of NK cell phenotypes. NK cells were gated as CD45+CD3âNK1.1+. For NK cell depletion, anti-mouse NK1.1 (PK136, BioXCell), which depletes NK cells73,74,75, or isotype control IgG2a (C1.18.4, BioXcell) were administered intraperitoneally at 8âmg per kg at 3 and 1 days before infection of 5- to 7-month-old mice.
For bulk RNA-seq analysis of mouse NK cells, purified NK cells from splenocytes of C57BL/6 mice were stimulated with IL-15 (50ângâmlâ1, BioLegend) for 4 days with or without fibrin. NK cells were stained with anti-CD3 (145-2C11, BD, 1:200), anti-NK1.1 (S17016D, BioLegend, 1:200), anti-CD45 (30-F11, BioLegend, 1:200) and aqua live/dead fixable dye on ice for 20âmin. The CD45+CD3âNK1.1+ live NK cells were sorted into 1.5âml tubes with 1âml of Buffer RLT Plus with 1% β-mercaptoethanol. RNA samples were prepared using the RNeasy Plus Micro Kit according to the manufacturerâs instructions. The cDNA library generation, quality control, sequencing and downstream analysis are performed as above.
Sample preparation for MS analysis
Human NK cells were isolated from freshly collected PBMCs (AllCells) using the NK cell Isolation Kit, Human (Miltenyi Biotec). In total, 5âÃâ106 NK cells were plated on each well of a six-well plate treated with or without fibrin for 1âh at 37â°C. Phosphoproteomic analysis was performed as described previously21,32. The samples were washed twice with cold PBS, lysed in 6âM guanidine hydrochloride (Sigma-Aldrich), then boiled at 95â°C for 5âmin, and stored on ice until sonication. Lysed samples were sonicated using a probe sonicator for 15âs at 10% amplitude, and protein was quantified by Bradford assay. Approximately 500âµg of protein sample was used for further processing, starting with reduction and alkylation using a 1:10 sample volume of tris-(2-carboxyethyl) (TCEP) (10âmM final) and 2-chloroacetamide (40âmM final) for 5âmin at 45â°C with shaking at 1,500ârpm. Before protein digestion, the 6âM guanidine hydrochloride was diluted sixfold with 100âmM Tris-HCL (pHâ8) to permit trypsin activity. Trypsin was then added at a 1:100 (w/w) enzyme:substrate ratio and placed in a thermomixer at 37â°C overnight (16âh) with shaking at 800ârpm. After digestion, 10% trifluoroacetic acid (TFA) was added to each sample to reach a final pH of 2. The samples were desalted using a vacuum manifold with 50âmg Sep Pak C18 cartridges (Waters). Each cartridge was activated with 1âml 80% acetonitrile/0.1% TFA, then equilibrated with 3âÃâ1âml of 0.1% TFA. After sample loading, the cartridges were washed with 3âÃâ1âml of 0.1% TFA, and the samples were eluted with 1âÃâ0.8âml 50% acetonitrile/0.25% formic acid. The samples were dried by vacuum centrifugation. The High-Select Fe-NTA phosphopeptide enrichment kit (Thermo Fisher Scientific) was used according to the manufacturerâs instructions with minor modifications for phosphopeptide enrichment. In brief, the samples were suspended in approximately one-third of the recommended binding/wash buffer volume (70âµl). After equilibrating the spin column, the resin slurry was resuspended in 210âµl of binding/wash buffer and divided into thirds. Each third of the resin was used for one sample. Tryptic peptides were mixed with the resin in a separate protein LoBind tube (Eppendorf) and incubated for 30âmin (at room temperature) on a thermomixer at 800ârpm. The samples were transferred on top of a 20âµl filtered tip, washed three times with binding/wash buffer and once with HPLC-grade water. The bound phosphopeptides were eluted with 70âµl elution buffer, and the pH was brought down immediately to nearly three with formic acid (10% (v/v) in HPLC-grade water). All of the samples were dried by vacuum centrifugation and stored at â80â°C until further analysis.
MS proteomics data acquisition
Dried phosphopeptides were resuspended in 0.1% (v/v) formic acid (Sigma Aldrich) in water (HPLC grade, Thermo Fisher Scientific) and analysed on the timsTOF HT mass spectrometer (Bruker Daltonics), paired with a Vanquish Neo ultra-high-pressure liquid chromatography system (Thermo Fisher Scientific). The samples were directly injected onto a PepSep C18 reverse-phase column (15âcm, 150âµm inner diameter, 100âà pore size, 1.5âµm particle size with UHP inlet, Bruker Daltonics) connected to a captive spray emitter (ZDV, 20âµm, Bruker Daltonics). Mobile phase A consisted of 0.1% (v/v) formic acid in water (HPLC grade, Thermo Fisher Scientific) and mobile phase B consisted of 0.1% (v/v) formic acid in 100% acetonitrile (HPLC grade, Thermo Fisher Scientific). Peptides were separated on a gradient from 3% to 25% mobile phase B over 47âmin, followed by an increase to 45% B over 8âmin, then to 95% over 1âmin, and held at 95% B for 4âmin for column washing at a flow rate of 200ânlâminâ1. Eluted peptides were ionized in a CaptiveSpray source (Bruker Daltonics) at 1,700âV. Raw data were acquired in data-independent acquisition coupled with parallel accumulationâserial fragmentation (dia-PASEF) mode with an optimized isolation window scheme in the m/z versus ion-mobility plane for phosphopeptides. The ion accumulation time and ramp times in the dual TIMS analyser were set to 100âms each. For dia-PASEF, in the ion mobility (1/K0) range 0.6 to 1.50âVsâcmâ2, the collision energy was linearly decreased from 59âeV at 1/K0â=â1.6âVsâcmâ2 to 20âeV at 1/K0â=â0.6âVsâcmâ2 to collect the MS/MS spectra in the mass range 400.2 to 1,399.3âDa. The estimated mean cycle time for the dia-PASEF windows was 1.38âs. The raw files were processed with Spectronaut (v.18.5, Biognosys) using its library-free DIA analysis with directDIA+ (Deep) search algorithm. Carbamidomethylation (cysteine) was set as a fixed modification for database search. Acetylation (protein N-term), oxidation (methionine), and phosphorylation (serine, threonine, tyrosine) were set as variable modifications. Reviewed human protein sequences (downloaded from UniProt, 6 October 2023) were used for spectral matching. The FDRs for the PSM, peptide and protein groups were set to 0.01, and the minimum localization threshold for PTM was set to zero. For MS2 level area-based quantification, the cross-run normalization option was unchecked (normalization was performed later using MSstats, see below), and the probability cut-off was set to zero for the PTM localization. We detected between 4,000 and 7,000 phosphorylated peptides per sample with an average percentage of phosphorylated to non-phosphorylated peptides of 73%.
Computational analysis of phosphoproteomics
Quantification of phosphorylation differences was performed using artMS as a wrapper around MSstats76, through functions artMS::doSiteConversion and artMS::artmsQuantification with the default settings. All peptides containing the same set of phosphorylated sites were grouped and quantified together into phosphorylation site groups. One sample outlier in intensity and peptide detection was discarded before quantitative analysis; unstimulated (mock) 1âh (PRIDE sample ID TOF01641_2_1_1683). For both phosphopeptide and protein abundance MSstats pipelines, MSstats performs normalization by median equalization, no imputation of missing values and median smoothing to combine intensities for multiple peptide ions or fragments into a single intensity for their protein or phosphorylation site group. Lastly, statistical tests of differences in intensity between infected and control timepoints were performed. When not explicitly indicated, we used the default settings for MSstats for adjusted P values. By default, MSstats uses the Studentâs t-tests for P value calculation and the BenjaminiâHochberg method of FDR estimation to adjust P values. Kinase activities were estimated using known kinaseâsubstrate relationships from the OmniPath database77. Kinase activities were inferred as a z-score calculated using the mean log2-transformed fold change of phosphorylated substrates for each kinase in terms of standard error (Zâ=â[Mâââμ]/s.e.), comparing fold changes in phosphosite measurements of the known substrates against the overall distribution of fold changes across the sample. To compare all phosphorylation sites across experimental groups as previously described32, a P value was also calculated from log2-transformed fold changes of all detected phosphorylation sites using a two-tailed Z-test method as shown in Fig. 3c, Extended Data Fig. 7b and Supplementary Tables 8â10. Network reconstruction and enrichment analysis of phosphoproteomics data were performed as described previously22.
Nanostring analysis
Formalin-fixed paraffin-embedded (FFPE) tissue was scrapped off into a 1.5âml Eppendorf tube and deparaffinized with 1âml of xylene for 2âmin and then pelleted and washed with 1âml of 100% ethanol. The samples were pelleted and incubated at room temperature until all of the residual ethanol had evaporated. Tissues were digested and RNA samples were isolated using the RNeasy FFPE Kit (Qiagen). The quantity was determined using the Nanodrop (Thermo Fisher Scientific) and the quality of RNA was determined on the Agilent Bioanalyzer. All of the samples passed quality control (>50% of RNA larger than 250 nucleotides). Gene expression assays were performed on the Nanostring nCounter machine with NS_Mm_HostResponse_v1.0 codeset. The raw data were processed and normalized counts, unadjusted P values and log2-transformed fold change values were generated with nSolver using two-tailed unpaired t-tests. For pathway analysis, the normalized counts of each gene were normalized across all of the samples to generate a z-score for heat maps of gene expression. The average z-score for each genotype was used for the heat map. Significantly downregulated genes between the 5B8 and IgG2b treated group (Pâ<â0.05) were on clusterProfiler to determine significantly downregulated pathways using the enrichGO function. The top 20 significantly downregulated pathways were used to generate the network.
Stereotactic injection of fibrinogen and spike
Fibrinogen was stereotactically injected into the brain as described previously35. Mice were anaesthetized with isoflurane and placed into a stereotaxic apparatus (Kopf Instruments). Alexa Fluor 488 human fibrinogen (Thermo Fisher Scientific) was dissolved in 0.1âM sodium bicarbonate (pHâ8.3) at 25â°C to 1.5âmgâmlâ1 (ref. 78), mixed with spike (4.6âmgâmlâ1), spike PVs (0.1âmgâmlâ1), bald PVs (0.1âmgâmlâ1) or PBS control (1:1 ratio), and incubated at 37â°C for 15âmin; 1.5âμl of the mixture was stereotactically injected at 0.3âμlâminâ1 with a 10âμl Hamilton syringe and a 33âgauge needle into the corpus callosum of 4- to 5-month-old C57BL/6 mice35. Mice were anaesthetized with avertin and transcardially perfused with 4% PFA in PBS. The brains were removed, post-fixed in 4% PFA overnight at 4â°C, processed with 30% sucrose, cut into 30âμm coronal sections and processed for immunohistochemistry. Images were acquired on the Axioplan II epifluorescence microscope (Zeiss) with Plan-Neofluar objectives (Ã10/0.3âNA). Images of similar anatomical locations were quantified using NIH ImageJ (v.1.50).
RNA in situ hybridization with immunohistochemistry
RNA in situ hybridization with immunohistochemistry was performed on brain sections from mice infected with Delta using RNAscope Multiplex Fluorescent Assay (ACD Bio) according to the manufacturerâs protocol for FFPE tissue. In brief, tissue was deparaffinized and incubated in 3% hydrogen peroxide for 10âmin, then subjected to antigen retrieval by boiling in RNAScope Target Retrieval Solution (ACD Bio) for 1âh. The samples were permeabilized with RNAScope Protease Plus reagent (ACD Bio) for 30âmin at 40â°C. RNA probes were hybridized to tissue for 2âh at 40â°C. Oligonucleotide probes for mouse Trem2, Cst7 and Spp1 were designed by ACD Bio (498711-C3, and 435191-C3, respectively). Probe signals were amplified using the RNAScope Multiplex Fluorescent Reagent Kit v2 (ACD Bio) and detected with TSA Vivid Fluorophore 570 (Tocris, 7526). Tissue sections were stained for one RNA probe and counterstained for IBA1 (234 308, Synaptic Systems, 1:500) using the RNA-Protein Co-Detection Ancillary Kit (ACD Bio). The slides were imaged using the Zeiss Axioplan 2 epifluorescent microscope at Ã20 and images were analysed using ImageJ (NIH). IBA1-postive microglia in each image were manually counted. Dense clusters of Trem2, Cst7 or Spp1 mRNA overlapping with IBA1 signal indicate microglia expressing disease-associated genes.
Statistical analysis
All values are reported as meanâ±âs.e.m. The ShapiroâWilk normality test79 was used to evaluate the normal distribution of the data. The equality of variance assumption was verified for both the responses in the natural and logarithmic scales using the BrownâForsythe test80. Comparisons between two matched-paired groups, where the assumption of normal distribution for the differences of paired responses was met, were performed using paired t-tests. P values for comparisons between two independent groups were calculated using MannâWhitney U-tests in the case of non-normally distributed data for which the equal variance assumption was not violated. For comparisons involving more than two groups, one- or two-way ANOVA followed by Tukeyâs post hoc test for multiple comparisons was used for data meeting normal distribution and equal variance assumptions. When the assumption of equal variance was violated, Welchâs t-tests were applied to log10-transformed response values, and the resulting raw P values were corrected for multiple testing using the Holm method57. For the survival analysis and weight change data, P values were calculated using the log-rank (MantelâCox) test and mixed-effects model, respectively. Sample sizes were determined by previous studies rather than statistical approaches. For all in vivo experiments, mice were randomized and experiments were conducted in a blinded manner to the mouse genotype, antibody or PV administration. Genotype and treatment assignment were revealed after image quantification. For bulk RNA-seq and Nanostring experiments, both mouse genotype and antibody treatment were blinded. SEM imaging and image acquisition were performed blinded to test conditions. Biochemical studies of the binding of fibrinogen to spike were performed in the Akassoglou laboratory and independently validated in the Greene laboratory and Assay Development and Drug Discovery Core with similar results.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.