Lentiviral constructs and production, in vitro transcription, cell lines and cell culture
All cells in this study were cultured in RPMI 1640 supplemented with 10% fetal bovine serum (FBS), 10âmM HEPES and 1% penicillin/streptomycin at 37â°C in fully humidified environment with 5% CO2 unless otherwise indicated. Cell lines were evaluated for mycoplasma contamination.
We used single chain variable fragment (scFv)-based CARs against human CD19 (clone FMC63) and the human TCR δ chain (clone 5A6.E9) with an appended N-terminal pentaglycine tag in a third generation lentiviral backbone (Extended Data Fig. 1). Both CAR constructs used scFvs in the light-chain-heavy-chain configuration followed by a CD8α hinge and transmembrane domain and CD137 and CD3ζ cytoplasmic domains. Furthermore, CARs were cloned with an IgG4 hinge instead of CD8α hinge domains (Extended Data Fig. 6c) with standard cloning methods.
To create a human CD19-sortase construct, we cloned human CD19 (C-terminal truncation) into a third generation lentiviral backbone that provided an IgH signal peptide followed by a flag-tag, low-affinity sortase mutant15 and a linker peptide. Similarly, we cloned a γδ TCR (clone PEER59) into this vector (both chains as a single transcript separated by a P2A site). Constructs were obtained as double-stranded DNA (dsDNA) fragments from IDT, digested and ligated into the lentiviral backbones using standard cloning techniques. We cloned extra TCR chains into lentiviral backbones (CD3γ-P2A-CD3δ and CD3ζ-P2A-CD3ε plasmids) to facilitate expression of the γδ TCR in non-T cell lines. Nucleotide and amino acid sequences of all constructs used in this study are listed in Supplementary Table 1. Lentivirus was produced as previously described using Lenti-X 293T cells60,61.
For in vitro messenger RNA (mRNA) transcription, CAR constructs were cloned into a pDrive.150 poly(A) backbone62 using standard cloning techniques and linear mRNA was transcribed using the T7 mScript Standard mRNA Production System (Cellscript). Linear dsDNA templates were generated by digesting with either ClaI or SpeI, and mRNA was synthesized following the manufacturerâs recommendations for a Cap 1-mRNA and roughly 150 base-long poly(A)-tail. mRNA was purified with an RNeasy Mini Kit (Qiagen), eluted in RNase-free water at 1âμgâμlâ1 and aliquoted and stored at â80â°C.
To disrupt the endogenous CD19 locus in Nalm6 cells (provided by M. Milone, originally obtained from DSMZ) and to create a cell line only expressing CD19-sortase or γδ TCR-sortase, two guide RNAs targeting the human CD19 locus were obtained (IDT, sequences in Supplementary Table 2) and 5âÃâ106 Nalm6 cells were electroporated with a total of 50âpM ribonucleoprotein (consisting of Cas9 (IDT) and single-guide RNA (sgRNA)) in a total volume of 20âμl of Lonza P3 buffer (P3 primary cell 4D-Nucleofector X kit S) with a Lonza 4D-Nucleofector Core Unit (pulse protocol EO115) according to the manufacturerâs protocol. After CD19 disruption, Nalm6 cells were cultured at 0.2â1âÃâ106âcells per ml in standard medium for 14âdays before sorting CD19 negative cells by FACS (BD Biosciences AriaII). Initial disruption efficiency was greater than 90%, which increased to more than 99% after sorting. CD19 negative Nalm6 cells were transduced with target proteins (CD3γ, CD3δ, CD3ζ, CD3ε, γδ TCR-sortase or CD19-sortase) and positive cells were enriched by FACS.
All target cells in this study express green fluorescent protein (GFP)-click beetle green luciferase. Target cells were sorted on a regular basis to ensure persistence of the luciferase and surface antigen expression over several passages.
Bulk and CD8 CART production
Bulk or CD8 only primary human T cells from healthy donors were obtained from the University of Pennsylvania Human Immunology Core, stimulated with anti-CD3/anti-CD28 beads (Dynabeads, Thermo Fisher) for 24âh before transduction with lentiviral CAR constructs. Anti-CD3/anti-CD28 magnetic beads were removed on day 4 after transduction and the IL-2 concentration was gradually lowered from 100 to 25âIUâmlâ1 by day 8 after activation and 0âIUâmlâ1 by day 10 after activation. Cell medium replacement and quantification of cell size and number (Coulter Multisizer 4e) occurred every 2â3âdays. CAR transduction efficiency was determined by flow using a polyclonal antimurine Fab antibody conjugated to biotin (Jackson ImmunoResearch) and streptavidin-PE. Non-transduced T cells from the same donor were stained under identical conditions and used as negative control.
To generate naive or effector CARTs, naive and effector T cells were isolated from bulk primary human T cells and were electroporated with mRNA encoding CARs. Naive T cells were isolated either with the Naive Pan T Cell Isolation Kit (Miltenyi Biotec, catalogue no. 130-097-095) or with a positive selection of CD62L and subsequent negative selection for CD45RA+ cells. For the latter approach, cells were stained with anti-CD62L-PE (BioLegend, DREG-56, catalogue no. 304840) and enriched with the anti-PE MultiSort kit (Miltenyi Biotec, catalogue no. 130-090-757) and LS column (Miltenyi Biotec, catalogue no. 130-042-401), with the flowthrough reserved for the isolation of effector T cells described below. CD62L+ cells were flushed out and separated from MultiSort MicroBeads using the MultiSort Release Reagent and centrifugation. CD45RA+CD62L+ cells were subsequently isolated by negative isolation using CD45RO MicroBeads (Miltenyi Biotec, catalogue no. 130-046-001) and two columns (Miltenyi LS). To isolate effector T cells of the same donor as the naive T cells, flowthrough from the first CD62L selection was added to the column (Miltenyi LD) for negative selection of CD62Lâ cells. More than 95% population purity (determined by flow cytometry) was used in the presented studies. Following isolation, naive and effector cells were electroporated with 10âμg mRNA/1âÃâ107 T cells encoding the CARs using Lonza 4D-Nucleofector Core Unit (pulse code EH115) according to the manufacturerâs protocol.
To disrupt the endogenous TCR, T cells were cotransduced with lentiviral CAR constructs and pCAT003, a lentivirus transfer plasmid encoding sgRNA targeting TRAC and gift from J. Doudna (Addgene plasmid no. 171628)63. Immediately following debeading, up to 4âÃâ106 T cells were electroporated with 50âpM of Cas9 as described above with the modification of pulse code EO115. TCRâ cells were negatively selected using CD3 MicroBeads (Miltenyi Biotec, catalogue no. 130-097-043) and LD column according to the manufacturerâs protocol.
To genetically disrupt IKZF1, 1âÃâ106 T cells were electroporated immediately following debeading with 50âpM Cas9 and 100ânM guide RNA (IDT, Supplementary Table 2) as described above, with the modification of pulse code EH115. Genomic DNA was isolated using DNeasy Blood & Tissue Kit (Qiagen, catalogue no. 69504) according to the manufacturerâs protocol. The targeted IKZF1 locus was amplified using indicated primers (Supplementary Table 3). Quantification of genetic editing efficiencies were estimated using tracking of indels by decomposition64. Western blots were performed and stained using rabbit antiIkaros (IKZF1) monoclonal antibody (Cell Signaling, 9034S; dilution 1:1,000), digital antirabbit-HRP (Kindle Biosciences, LLC, R1006; dilution 1:1,000), mouse anti-β-actin monoclonal antibody (Cell Signaling, 3700S; dilution 1:3,000) and digital antimouse-HRP (Kindle Biosciences, LLC, R1005; dilution 1:3,000). Uncropped images of blots are provided in Supplementary Fig. 1. Pharmacologic depletion of IKZF1 was performed using 0.1âμM lenalidomide (MedChemExpress, catalogue no. HY-A0003).
LIPSTIC assay
Biotinylated LPETG peptide (biotin-aminohexanoic acid–LPETGS, C-terminal amide, 95% purity)15 was purchased from LifeTein (custom synthesis), reconstituted in PBS at 10âmM and stored at â80â°C.
To label target cells, Nalm6 cells (expressing sortase-tethered target molecules) were incubated with biotinylated LPETG peptide (100âμM, LifeTein) for 30âmin at 37â°C in RPMI/10%FBS, followed by washing three times to remove excess soluble peptide. Sortase-bound LPETG was then labelled with fluorescent streptavidin (PE, AF647 or APC; 10âμgâmlâ1; BioLegend) for 30âmin at 37â°C. Cells were then washed three times and resuspended at 1âÃâ106 cells per ml.
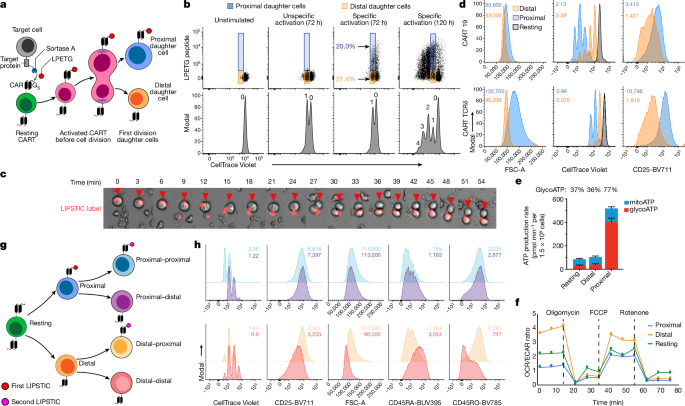
All LIPSTIC assays were performed using fully rested T cells that had not demonstrated cell number increases in roughly 2âdays. For the CARTs in the presented studies, the transduction efficiencies were between 20 and 85%, and the cell sizes of rested T cells were between 200 and 260âfl, which was achieved 12â15âdays after activation. T cells were stained with CellTrace Violet following the manufacturerâs recommendations with the following modifications: T cells were stained at a concentration of 1âÃâ107âcells per ml for 10âmin at 37â°C. Target cells and CARTs were mixed in a six-well plate well in a total volume of 5.5â7âml, and CART to target ratios ranged from 0.3:1 to 4.25:1. Cells were incubated for 72âh before cell sorting (BD Biosciences AriaII) and subsequent analysis of first-division daughter cells.
A second target encounter LIPSTIC assay was performed using sorted first-division LPETG+ or LPETGâ cells. Target cells labelled with a second colour fluorescent streptavidin and CARTs were mixed in a 96-well plate in a total volume of 200âμl and a 1:1 CART:target ratio. Second-division daughter cells were sorted 24âh after coincubation.
Sorting gates were established for live single cells that were negative for GFP (excluding target cells), positive for Cell Trace Violet (CTV) and positive or negative for LPETG. LPETG positivity was determined relative to untransduced T cells, CARTs incubated without target cells or irrelevant CARTs incubated with target cells (threshold for LPETG positivity was generally the same for all controls).
Multiparametric flow cytometry analysis of T cells
Unless otherwise specified, antibodies were purchased from BioLegend.
In vitro and in vivo LIPSTIC assay populations were sorted and subsequently phenotyped by staining with 1:200 CD8-APCH7 (SK1, BD Biosciences, catalogue no. 561423), 5:400 CD4-BUV805 (SK3, BD Biosciences, catalogue no. 612887), 1:160 CD45RA-BUV395 (HI100, BD Biosciences, catalogue no. 740298), 5:400 CD45RO (UCHL1, catalogue no. 304234), 1:40 CD25-BV711 (M-A251, catalogue no. 356138) and/or 1:250 CD62L-PE (DREG-56, catalogue no. 304840).
For in vivo studies, samples were stained with CD8-APC/Cy5.5 (RFT8, SouthernBiotech, catalogue no. 9536-18), CD4-PE/Cy5.5 (RFT4, SouthernBiotech, catalogue no. 9522-16), CD3-BV605 (OKT3, catalogue no. 317322), CD19-APC (HIB19, catalogue no. 302212), CD45RA-BUV395 (HI100, catalogue no. 740298), CD45RO-BV785 (UCHL1, catalogue no. 304234), CD62L-PE (DREG-56, catalogue no. 304840), TCR-alpha/beta-BV421 (IP26, catalogue no. 306722) and CD45-PECy7 (QA17A19, catalogue no. 393408) at 1:100 dilution. Whole blood was stained in Trucount tubes (BD Biosciences) and fixed with FacsLyse solution (BD Biosciences) according to the manufacturerâs recommendations. Single-cell suspensions from spleen samples were produced by homogenization of the tissue through a 70âμm mesh followed by treatment with Red Blood Cell Lysis Buffer (BioLegend) according to the manufacturerâs recommendations and stained in PBS, 1% FBS and cell numbers were quantified with CountBright Plus Absolute Counting Beads (Thermo Fisher).
Samples were analysed on an LSRII, LSR Fortessa or FACSymphony A3 Cell Analyzer (BD Biosciences). The population of interest was gated based on forward- versus side-scatter characteristics followed by singlet gating. Data were analysed with FlowJo v.10 (Tree Star). Graphs and statistical analyses were generated using GraphPad Prism v.9.4.0.
Live-cell microscopy
For live-cell imaging to capture CARTs undergoing the first cell division, LPETG-positive CARTs before the first cell division were isolated using fluorescence activated cell sorting after 48âh of coincubation with target cells and then imaged in a humidified incubation chamber at 37â°C in 5% CO2 on a Zeiss Observer 7 equipped with a Zeiss Axiocam 702 monochrome CMOS camera, a Zeiss Axiocam 503 colour CCD camera and a Colibri 7 LED light source in Definite Focus mode in a 35âmm glass bottom dish (Ibidi) every 3âmin.
To image the transfer of LPETG peptide from target to CART cells, CTV-labelled T cells were incubated with LPETG peptide-loaded target cells at an effector to target cell ratio of 1:5. The excess of target cells in this context increases the frequency of observing the interaction between CARTs and targets. Cells were placed in a 35âmm glass bottom dish (Ibidi) and analysed in a humidified incubation chamber at 37â°C in 5% CO2. Photographs were acquired in the GFP, CTV and AF647 channels in Definite Focus mode on a Zeiss Observer 7 every 45âs for 50âmin. Images were acquired with Ã40 objective using Zen (Blue edition) software (v.2.5, Zeiss). Videos were created with Fiji-ImageJ.
Metabolic analysis
T cell metabolic profiles were assessed using the Seahorse mitochondrial stress test (Agilent Technologies). Individual wells of an XF96 cell-culture microplate were coated with CellTak as per the manufacturerâs instructions. The matrix was adsorbed overnight at 37â°C, aspirated, air-dried and stored at 4â°C until use. Mitochondrial function was assessed on day 0 or day 1 after sorting proximal or distal or undivided cells. T cells were resuspended in non-buffered RPMI 1640 medium containing 5.5âmM glucose, 2âmM l-glutamine and 1âmM sodium pyruvate and seeded at 1.5âÃâ105 cells per well. The microplate was centrifuged at 1,000g for 5âmin and incubated in standard culture conditions for 60âmin. During instrument calibration (30âmin), the cells were switched to a CO2-free 37â°C incubator. XF96 assay cartridges were calibrated according to the manufacturerâs instructions. Cellular OCRs and ECARs were measured under basal conditions and following treatment with 1.5âμM oligomycin, 1.5âμM FCCP and 40ânM rotenone, with 1âμM antimycin A (XF Cell Mito Stress kit, Agilent). OCR/ECAR ratios are calculated using the mean OCR and ECAR of 3â5 replicates for each population.
In vivo studies
Immunodeficient NSG (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ) mice were bred in house under an approved Institutional Animal Care and Use Committee (IACUC) protocol and maintained under pathogen-free conditions. To facilitate engraftment of T cells, bulk (CD4+ and CD8+) T cells were used in in vivo studies19. Sample sizes were not predetermined based on statistical methods but were chosen based on preliminary data and previously published results. For all in vivo experiments, treatment groups were randomly selected by the cage number. In vivo injections were performed in a blinded fashion by a member of the Ellebrecht or Payne laboratories or a staff member of the Human Stem Cell and Xenograft Core of the University of Pennsylvania.
For in vivo LIPSTIC, LPETG-labelled target cells were injected intraperitoneally immediately followed by CTV-labelled CARTs. A 1:1 target:CART ratio was maintained, with a total of 1âÃâ107 total T cells injected. Mice were euthanized 2âdays following injection. Cells were collected using peritoneal wash three times using 5âml of ice-cold 2% FBS and PBS. First-division daughter cells were sorted and assessed by flow cytometry as described above.
For functional longevity studies, 2.5âÃâ105 anti-CD19 CAR proximal- or distal-daughter cells, non-activated resting anti-CD19 CARTs or non-transduced T cells were intravenously injected into NSG mice on day 0. After 35âdays, mice were challenged with 1âÃâ106 Nalm6 cells. Leukaemia burden was determined by bioluminescence imaging. Bioluminescence was quantified with an IVIS Lumina III (PerkinElmer) 2â3 times per week after Nalm6 injection as previously described3.
In the stress-test model, NSG mice were injected with 1âÃâ106 Nalm6 cells on day 0. Engraftment of Nalm6 was confirmed on day 3 by bioluminescence imaging. On day 4, mice were treated with 2.5âÃâ105 proximal or distal-daughter anti-CD19 CARTs; 2.5âÃâ106 non-activated resting anti-CD19 CARTs or non-transduced T cells; or 1.3âÃâ106 bulk restimulated anti-CD19 CARTs (unsorted) by intravenous injection. Leukaemia burden was determined with bioluminescence imaging as above. Mice were euthanized when they had reached a total bioluminescence flux of at least 1âÃâ1011 photons per second, demonstrating loss of leukaemia control.
Peripheral blood was obtained by retro-orbital bleeding. Mice were euthanized for organ harvest according to local IACUC guidelines, and spleen and blood samples were assessed by flow cytometry as described above. In accordance with the approved IACUC protocol for these studies, humane endpoints for euthanizing mice were established and not exceeded in this study. Mice were monitored at least twice weekly for symptoms. If severe lethargy or weakness, hunching, emaciated body condition (body condition score of 1 out of 5) or loss of 20% or more body weight, were observed, mice were euthanized. If a body condition score of 2 out of 5 was observed and accompanied by lethargy, mice were euthanized. Furthermore, mice were euthanized when their total bioluminescence flux exceeded 1âÃâ1011. All studies involving animals were performed under a protocol approved by the University of Pennsylvania IACUC.
Luciferase-based in vitro cytotoxicity assay
Cytotoxicity assays were performed either on day 1 or 4 after first cell division as previously described3. Click beetle green luciferase-expressing target Nalm6 cells were cocultured with proximal, distal or resting CARTs or donor-matched non-transduced T cells at indicated E:T ratios. To test TCR-mediated cytotoxicity of mRNA-electroporated naive and effector CARTs, K562 cells positive for CD64 (FcγRI) were incubated with 100âμgâmlâ1 anti-human CD3 (OKT3, Invitrogen, catalogue no. 16-0037-85) for 30âmin on ice and washed twice were used as target cells.
Single-cell multiomic analysis
First-division proximal, first-division distal, activated-undivided and resting CD8 CARTs (1.5âÃâ105 cells each), sorted as described above from the LIPSTIC assay, were separately incubated in flow cytometry staining buffer (BioLegend) with a custom TotalSeq-C antibody cocktail (Supplementary Table 4, BioLegend 900000114, lot B311489) in 100âμl for 30âmin at 4â°C before washing three times. Cell concentration was adjusted to 1.5âÃâ106 live cells per ml. 10,000 live CD8 T cells from each LIPSTIC population were loaded onto NextGem K chips (10X Genomics) and processed in a 10X Chromium device according to the manufacturerâs recommendations. A biological replicate was performed with CART from a separate donor. Library preparation was performed according to the 10âÃâ5â² V2 protocol for antibody-derived tags (ADT), gene expression (GEX) and paired alpha and beta TCR chains. Complementary DNA and subsequent library intermediates were checked for correct size, appropriate quantity and quality with a DNA high-sensitivity kit on a Bioanalyzer 2100 (Agilent). Libraries were sequenced in paired-end dual-index mode for 150âÃâ2 cycles on a NovaSeq 6000 sequencer (Illumina, one lane of a S4 cartridge). All cells in each experiment were sorted and stained on the same day and libraries were processed in parallel and sequenced in the same lane to minimize batch effects. Counts for demultiplexed GEX, ADT and TCR libraries were obtained with the STAR method of the Cell Ranger multi pipeline (10X Genomics, Cell Ranger v.6.1.2) using the human GENCODE v.32/Ensembl 98 GRCh38 reference (detailed version by 10X Genomics: Human (GRCh38) 2020-A, Human (GRCh38) v.5.0.0), which then were aggregated with the Cell Ranger aggr pipeline with read depth normalization to further reduce batch effects across libraries. Downstream analysis was performed with the Seurat V4 R package22. To remove doublets and low cells, cells with more than 25% mitochondrial gene transcripts, less than 7.5% ribosomal gene transcripts, transcript counts less than 500 or greater than 40,000, or a minimum number of detected genes of less than 500 were excluded. Counts were single-cell transformed using the sctransform V2 and glmGamPoi packages65. Dimensionality reduction was performed based on ADT counts with subsequent analysis of genes and surface proteins of interest and differentially expressed genes or surface proteins for TN-, TCM-, TEM– and TRM-like subsets. RNA velocity analysis was performed by counting spliced and unspliced transcripts in Cell Ranger binary alignment map output files with the velocyto package31 using the same transcriptome reference gene transfer format file (refdata-gex-GRCh38-2020-A) that was used for the initial Cell Ranger run. Output loom files were then used in scvelo after export of TN, TCM, TEM and TRM expression matrices containing proximal and distal first-division daughter cells from Seurat and conversion to SCANPY/ANNDATA objects66. Global velocity vectors and velocities of genes of interest were computed and visualized in stochastic or dynamic mode with scvelo30. Regulon analysis (gene modules co-expressed with transcription factors and with correct cis-regulatory upstream motif for a respective transcription factor) was performed using a list of 1,390 human transcription factors (https://github.com/aertslab/pySCENIC/blob/master/resources/hs_hgnc_curated_tfs.txt) with the Python version of SCENIC (that is, pySCENIC v.0.11.2)32,67 after importing expression matrices from Seurat to SCANPY (v.1.7.2). Gene-set enrichment analysis68,69 was performed on each T cell subsets with the GSEA function of clusterProfiler70 in R. T cell clonal analysis was performed with scRepertoire71 in R using the âstrictâ setting, which requires identical V, D (if applicable), J and C genes in addition to identical CDR3 nucleotide sequence of both TCR chains to identify T cells belonging to the same clonotype.
Seurat analysis was performed in R v.4.3.1, velocity analysis was performed in Python v.3.10.4 and regulon analysis was performed in Python v.3.7.12 in accordance with the respective pipeline requirements.
Statistical analysis
Statistical significance was determined with two-sided tests unless otherwise indicated. Where appropriate and as indicated, P values were adjusted for multiple testing (BenjaminiâHochberg). Q values were calculated with clusterprofiler in R. Whenever individual data points are presented, a horizontal line represents the mean value of the group. Survival in in vivo experiments was defined as time until the predetermined bioluminescence threshold was reached. KaplanâMeier statistical analysis was used to compare survival between groups. Unless otherwise indicated, asterisks depict the following significance values: *Pâ<â0.05, **Pâ<â0.01, ***Pâ<â0.001. P values less than 0.05 were considered statistically significant. The mean fluorescence intensity in flow cytometry plots is labelled with a cross in each gate. Statistical analysis was performed with the respective pipelines as mentioned above or with GraphPad Prism v.9.4.0.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.