Animal experiments
All mouse experiments were performed in accordance with institutional guidelines for animal care and use established in the Principles of Laboratory Animal Care (directive 86/609/EEC). Animal work was only initiated upon approval by the Italian Istituto Superiore di Sanità (ISS) with authorization no. 40/2022, protocol 22418.167 or by the Animal Ethics Committee of the Netherlands Cancer Institute. Six-to-eight-week-old female athymic nude mice (purchased from Envigo) were used in the experiments at the Animal Facility of the IRCCS Ospedale Policlinico San Martino (Genoa). These mice were housed in Sealsafe Plus GM500 individually ventilated cages (IVCs) held on DGM Racks at 22 ± 2 °C and approximately 50–60% relative humidity under a 12 h:12 h light:dark lighting cycle and with food (standard diet, 4RF18, Mucedola) and water ad libitum. Mice were acclimatized for one week before experiments were initiated. To allow MCF7 xenograft growth, a 17β-oestradiol-releasing pellet (Innovative Research of America) was inserted in the intra-scapular subcutaneous region under anaesthesia conditions, the day before cell injection. Xenografts were established by subcutaneous injection of 5 × 106 MCF7 cells to both flanks of the mouse (experiments in Figs. 1a,b, 2 and 4a,b), or orthotopic injection of 3 × 106 MCF7 cells into the fourth abdominal fat pad (experiments in Fig. 4d and Extended Data Fig. 3a,b). Treatment was initiated when the tumours appeared as established palpable masses (~2 weeks after cell injection). In each experiment, mice were randomly assigned to receive one of the following treatments or their combinations, as indicated: control (ad libitum diet); TMX (45 mg kg−1 per day in peanut oil, oral gavage3,39), fasting (water only, for 48 h every week3,40), Dexa (4 mg kg−1 every other day in physiological solution, intraperitoneal41), IGF1 (200 μg kg−1 body weight, intraperitoneal twice a day on the days of fasting); insulin (20 mU kg−1 body weight, intraperitoneal once a day on the days of fasting); leptin (1 mg kg−1 body weight, intraperitoneal once a day on the day of fasting). During the 48 h of fasting, mice were individually housed in a clean, new cage to reduce coprophagy and the intake of the residual chow. Body weight was measured immediately before, during and after fasting. Fasting cycles were repeated every seven days to allow for complete recovery of body weight before a new cycle. The size of the tumours was measured twice a week and tumour volume was calculated using the formula: tumour volume (in mm3) = (w2 × W) × π/6, where w and W are lengths of the minor side and major side (in mm), respectively. The maximum tumour volume that was permitted by our Institutional Animal Care and Use Committee (IACUC) was 1,500 mm3, and in none of the experiments were these limits exceeded. Tumour masses were isolated at the end of the last fasting cycle, weighed, divided into two parts, snap frozen in liquid nitrogen and stored at −80 °C. Ten slices of 50 μm per tumour sample were subsequently utilized for ChIP–seq, proteomics and RNA-seq analyses. Sample size estimation was performed using PS (power and sample size calculation) software (Vanderbilt University). By this approach, we estimated that the number of mice that were assigned to each treatment group would reach a power of 0.85. The type I error probability associated with our tests of the null hypothesis was 0.05. Mice were assigned to the different experimental groups in a random fashion. Operators were unblinded, as blinding during animal experiments was not possible because mice were subject to a specific diet supply and daily treatment.
To establish mammary intraductal cell line-derived xenograft (MIND-CDX) models, T47D cells were intraductally injected as previously described39,42. Specifically, 1 × 106 T47D cells were dissociated to single cells with 0.05% trypsin and injected intraductally into the abdominal/inguinal mammary glands (both sides) of 8-week-old female NSG mice (Jackson Laboratory) with a 34G needle. To ensure stable outgrowth, T47D MIND-CDX mice were supplemented with 17β-oestradiol (Sigma, E2758) via the drinking water at a concentration of 4 µg ml−1 starting 7 days prior to tumour inoculation via intraductal injection. E2 supplementation was maintained throughout the experiment. To establish TSAE1 allograft models, BALB/c mice (Jackson Laboratory) were intraductally injected with of 1 × 104 single cells in PBS as described above (one gland). The IDC186 MIND-PDX model was established from a pre-menopausal Caucasian breast cancer patient, confirmed positive for ERα (95%) and PR (95%) but negative for HER2 (Extended Data Fig. 6h). To generate the PDX model, 5 × 104 single cells in PBS were intraductally injected into one of the abdominal mammary glands of 8-week-old female NSG mice (Jackson Laboratory) and supplemented with 17β-oestradiol (Sigma, E2758) as described above.
The xenograft model cohorts were monitored three times per week and tumours were palpated and measured via calliper in two dimensions. Mice were enrolled into treatment groups when the largest tumour per animal measured 50 mm3 for T47D and IDC186 xenografts and 25 mm3 for TSAE1 allografts, respectively. Mice were randomly allocated into treatment groups and received the following treatments: (1) vehicle treatment (corn oil, daily, oral gavage); (2) TMX (45 mg kg−1 in corn oil, daily, oral gavage); (3) Dexa (4 mg kg−1, 3 times per week, intraperitoneal injection); or (4) TMX plus Dexa. Mice were treated for 28 consecutive days for the TSAE1 and IDC186, 56 days for T47D (with a 1-week treatment break between days 28 and 35), or until the cumulative mammary tumour burden reached a volume of 1,500 mm3 and thus the maximally permitted disease end point. At euthanasia, mammary glands and full female reproductive tracts were collected in formalin, stained against haematoxylin and eosin (H&E) according to routine procedures, and uteri were analysed for histopathological abnormalities. Tumour measurements and post-mortem analysis were performed in blinded fashion. H&E slides were reviewed by a trained pathologist (J.-Y.S.) in a blinded manner. Slides were digitally processed using a PANNORAMIC 1000 whole slide scanner (3DHISTECH) and captured with the Slidescore software (www.slidescore.com).
Clinical studies of FMD in patients undergoing endocrine therapy for HR+ breast cancer
The NCT05748704 trial was conducted at the IRCCS Ospedale Policlinico San Martino (Genoa), between December 2022 and February 2024 and was approved by the Comitato Etico Regione Liguria. This trial consists of a single-arm phase I/II clinical study of a FMD with solid tumours who are candidates to receive active medical or radiotherapy treatment (or with medical treatment or radiotherapy already ongoing). The nutritional intervention consists of a low-calorie diet lasting 5 days and aimed at providing between 800 and 1,000 kcal day−1 (tentatively 10% carbohydrates, 15% proteins and 75% lipids). Throughout the clinical study, patients have received dietary counselling for the intervals between FMD cycles, aiming at providing an appropriate intake of proteins, essential fatty acids, vitamins and minerals43 and have also been invited to perform light/ or moderate daily muscle training to enhance muscle anabolism44. Study primary outcomes were the effects of the FMD regimen on the circulating levels of factors with pro- or anti-oncogenic activity (including insulin, IGF1, IGFBP1, IGFBP3, leptin, adiponectin, IL-6, TNF and IL-1β), as well as the effect of FMD cycles on leukocyte subpopulations with a role in tumour growth control, such as regulatory T cells, myeloid-derived suppressor cells (MDSCs) as well as natural killer (NK) cells, and its stem cell pool (for example, haematopoietic stem cells, endothelial stem cells, mesenchymal stem cells). Additional information on this trial is available at https://clinicaltrials.gov/ct2/show/NCT05748704. Patient serum for subsequent ELISA assays of circulating growth factors and adipokines has been routinely collected before and after the first, sixth and twelfth FMD cycle. Informed consent was obtained from all patients participating in the clinical trial.
The DigesT study (ClinicalTrials.gov ID: NCT03454282) trial was conducted between July 2018 and December 2020 and in accordance with the Declaration of Helsinki and the principles of Good Clinical Practice. The study protocol was approved by the Institutional Review Board (IRB) and the Ethics Committee of Fondazione IRCCS Istituto Nazionale dei Tumori Milan (INT 157/17). All patients provided written informed consent before any study procedures, as well as for the use of clinical and biological data for research purpose. The FMD nutritional intervention consisted in a 5-day, plant-based, calorie-restricted (up to 600 kcal on day 1; up to 300 kcal on days 2, 3, 4 and 5), low-carbohydrate, low-protein, nutritional regimen, as previously published6. Enrolled patients initiated the FMD 12–15 days before surgery, and underwent blood sampling after at least 8 h complete fasting on the morning (08:30 to 10:00) of FMD initiation (pre-FMD), and on the morning of FMD completion (post-FMD). Tumour samples for RNA-seq analyses were obtained diagnostic core biopsies performed at baseline (Pre-) and from matched surgical specimens (Post-). The primary outcomes of the study were to measure the absolute and relative changes in population of peripheral blood mononuclear cells before and after the FMD. Additional information on this trial is available at https://clinicaltrials.gov/ct2/show/NCT03454282.
ChIP–seq
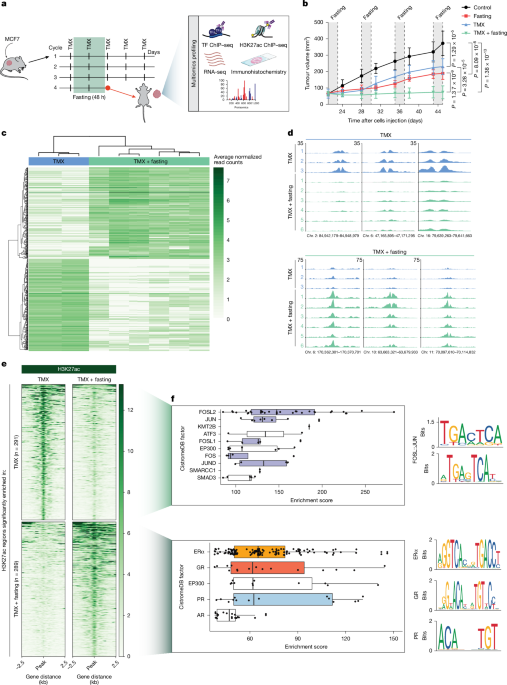
Snap-frozen xenografted tumours were double fixed using 2 mM of disuccinimidyl glutarate diluted in solution A (50 mM Hepes, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA) for 25 min followed by 1% formaldehyde for 20 min, at room temperature. Cells were then lysed and sonicated accordingly to the protocol previously described45, with the difference of have performed 15 cycles of 30 s on, 30 s off in the sonication step (BioRuptor Pico, Diagenode). Obtained nuclear lysates were incubated overnight with 50 μl of protein A coated Dynabeads magnetic beads (10008D, Invitrogen) conjugated with 5 μg of ERα (06-935, Millipore), H3K27ac (39133, Active Motif), GR (12041S, Cell Signaling), PR (8757S, Cell Signaling) or JUN (9165S, Cell Signaling) antibodies. The resulting immunoprecipitated DNA was submitted for library preparation using the KAPA library kit (KK8234, Roche) and subsequently paired-end sequenced on the Illumina NovaSeq 6000 system with read length of 51 bp. ChIP–seq analyses were performed using an in house pipeline publicly available at https://github.com/sebastian-gregoricchio/ChIP_Zwart (v.0.1.2) with default parameters. In brief, all samples were aligned to reference genome Hg38/GRCh38 using Burrows-Wheeler Aligner46 (BWA v.0.5.10). Reads were filtered based on mapping quality (MAPQ ≥ 20), and duplicate reads were marked with Picard MarkDuplicates (v.2.19.0). MACS2 (v.2.1.2) was used to perform peak calling over input ChIP–seq samples; only peaks with a q-value < 0.01 were retained. DeepTools47 (v.2.5.3) was used to calculate the fraction of reads in peaks (FRiP) and normalized ChIP–seq signal. For visualization purposes, Reads Per Genomic Content (RPGC) normalization (1× coverage) signal was averaged among the replicates per each condition using deeptools bigwigCompare. Genome browser snapshots were generated using the R v.4.0.3 environment and Rseb48 (v.0.3.1) (https://github.com/sebastian-gregoricchio/Rseb). Tornado plots were generated using deepTools (v.2.5.3). Differential peak analyses were performed using diffBind49 (v.3.0.15). Peaks were defined as differential when the |log2(fold change)| >1.5 and adjusted P value <0.05. Genomic location annotation of the peaks was performed using ChIPSeeker50 (v.1.26.2) defining the promoter region as −2 kb:transcription start site:+1 kb. Transcription factor binding enrichment from public available datasets—GIGGLE analyses51—were performed using the tool available at the of CistromeDB website (http://cistrome.org/db/).
Cell lines
The MCF7, T47D and HEK293T cell lines were purchased from the American Type Culture Collection (ATCC). TSAE1 cells were a gift from C. Isacke laboratory28. All cell lines were kept in DMEM, high glucose, pyruvate (Gibco) and supplemented with 10% fetal bovine serum (FBS, Capricorn Scientific) and 1% penicillin-streptomycin (5,000 U ml−1, Life Technologies). For ligand treatment, 4-hydroxytamoxifen (HY16950; MedChemExpress) and SR11302 (HY-15870; MedChemExpress) were reconstituted in DMSO, and used in the described concentrations and time points. All cell lines were cultured at 5% CO2 at 37 °C, were subjected to regular Mycoplasma testing, and underwent authentication by short tandem repeat profiling (Eurofins Genomics).
CRISPR–Cas9-mediated knockout cell line generation
GR-targeting single-guide RNA (NR3C1; ATGACTACGCTCAACATGTT) and non-targeting (NT) control guide RNA (GTATTACTGATATTGGTGGG) were separately cloned into the lentiCRISPR v.2 vector52. Using H3K293T cells, the CRISPR vectors were co-transfected with third-generation viral vectors using polyethyleneimine (PEI, Polysciences). After lentivirus production, the medium was collected and added to the MCF7 cells. Two days after infection, cells were selected for 2 weeks with 2 μg ml−1 puromycin (Sigma Aldrich), and knockout efficiency was confirmed by western blot and immunofluorescence.
Immunoblotting
Total protein lysates were obtained using Laemmli buffer complemented with 1× complete protease inhibitor cocktail (Roche) and 1× phenylmethylsulfonyl fluoride (PMSF). Forty micrograms of protein per sample was resolved in a NuPAGE 4–12% Bis-Tris gel (NP0335BOX, Invitrogen) in 1× NuPAGE MOPS SDS Running Buffer (NP00012, Invitrogen) and sequentially transferred to a 0.45-μm nitrocellulose membrane (Santa Cruz Biotechnology). Protein detection was performed using antibodies raised to detect GR (1:1,000, 12041S, Cell Signaling) and β-actin (1:10,000, MAB1501R, Merck Millipore). Odyssey CLx Imaging system (Li-Cor Biosciences) and ImageStudio Lite v.5.2.5 (LI-COR Biosciences) software were used to scan and visualize the proteins.
Immunofluorescence
Cells were fixed with 2% paraformaldehyde (103999, Merck) for 10 min, washed twice with PBS and subsequently permeabilized with 0.5% Triton/PBS (Triton X-100, Sigma Aldrich). Following two PBS washing steps, cells were blocked for 1 h in 1% bovine serum albumin (BSA, A8022, Sigma/Merck)/PBS solution before incubation with antibody against GR (1:100, 12041S, Cell Signaling). After two additional PBS washes, cells were incubated with Alexa Fluor 488 goat anti-rabbit IgG (1:1,000, A-11008, ThermoFisher Scientific) and DAPI (ProLong Gold Antifade Mountant, P36930, ThermoFisher Scientific) and signal detected using laser confocal microscopy (SP5, Leica).
Immunohistochemistry
Immunohistochemistry of the formalin-fixed, paraffin-embedded (FFPE) tumour samples was performed on a BenchMark Ultra (Ki-67) or a Discovery Ultra (ERα, PR, GR) automated stainer (Ventana Medical Systems). In brief, paraffin sections were cut at 3 µm, heated at 75 °C for 28 min and deparaffinised in the instrument with EZ prep solution (Ventana Medical Systems). Heat-induced antigen retrieval was carried out using Cell Conditioning 1 (CC1, Ventana Medical Systems) for 32 min (Ki-67, ERα, PR) or 64 min (GR) at 95 °C. Ki-67 was detected using the clone 30-9 (Ready-to-Use, 32 min at 37 °C, Roche Diagnostics/Ventana), GR using the clone D6H2L (1/600 dilution, 1 h at 37 °C, 12041, Cell Signalling), ERα using the clone SP1 (Ready-to-Use, 32 min at room temperature, Roche Diagnostics/Ventana) and PR using the clone 1E2 (Ready-to-Use, 32 min at room temperature, Roche Diagnostics/Ventana). In order to reduce background signal for the PR staining, after primary antibody incubation slides were incubated with normal antibody diluent (Roche Diagnostics) for 24 min. Bound Ki-67 antibody was detected using the OptiView DAB Detection Kit (Ventana Medical Systems). GR and ERα bound antibody was visualized using Anti-Rabbit HQ (Ventana Medical systems) for 12 min at 37 °C followed by Anti-HQ HRP (Ventana Medical systems) for 12 min at 37 °C and the ChromoMap DAB detection kit (Ventana Medical Systems). PR bound antibody was detected using OmniMap anti-Rabbit HRP (Ventana Medical systems) for 12 min at room temperature. followed by ChromoMap DAB detection kit (Ventana Medical Systems). Slides were counterstained with Hematoxylin and Bluing Reagent (Ventana Medical Systems). A PANNORAMIC 1000 scanner from 3DHISTECH was used to scan the slides at a 40× magnification and uploaded to the Slidescore software (www.slidescore.com). Digitized slides were further processed using QuPath (v.0.6.0)53. The analysis protocol begins with tissue detection using a pixel classifier, which differentiates foreground from background (white) pixels. Next, manual annotation is performed with the brush tool to delineate and exclude stroma areas from the region of interest. To identify Ki-67-positive and negative cells, the native cell detection tool is used with the optical density sum option for image analysis. Default parameters are applied, with adjustments made only to the pixel size (0.25 µm), based on the slide resolution, and the target cell size (8 µm) for accurate cell identification. Finally, the scoring is calculated as the proportion of positive cells relative to the total number of detected cells, providing a quantitative assessment of Ki-67 expression. All quantifications were compared and approved by a trained pathologist (J.S.).
RNA-seq analyses
MCF7 xenograft tumour RNA was isolated by homogenizing the tissue sample in 1 ml of RLT buffer (79216, Qiagen) and 1% β-mercaptoethanol using the Qiagen TissueLyserII (85300, Qiagen) for 6 min with frequency setting 30 (1 s−1) in combination with 5-mm stainless steel beads (69989, Qiagen). The total RNA was isolated using the RNeasy Mini Kit (74106, Qiagen), including an on column Dnase digestion (79254, Qiagen), according to the manufacturer’s instructions (Rneasy Mini Handbook, Qiagen). Quality and quantity of the total RNA was assessed by the 2100 Bioanalyzer using a Nano chip (Agilent). Total RNA samples having RNA integrity number (RIN) >8 were subjected to library generation. Strand-specific libraries were generated using the TruSeq Stranded mRNA sample preparation kit (Illumina, RS-122-2101/2) according to the manufacturer’s instructions (Illumina, document 1000000040498 v.00). In brief, polyadenylated RNA from intact total RNA was purified using oligo-dT beads. Following purification the RNA was fragmented, random primed and reverse transcribed using SuperScript II Reverse Transcriptase (Invitrogen, 18064-014) with the addition of actinomycin D. Second strand synthesis was performed using polymerase I and RnaseH with replacement of dTTP for dUTP. The generated cDNA fragments were 3′ end adenylated and ligated to IDT xGen UDI(10 bp)-UMI(9 bp) paired-end sequencing adapters (Integrated DNA Technologies) and subsequently amplified by 12 cycles of PCR. The libraries were analysed on a 2100 Bioanalyzer using a 7500 chip (Agilent), diluted and pooled equimolar into a multiplex sequencing pool. The libraries were sequenced with 51 paired-end reads on a NovaSeq 6000 using a Reagent Kit v.1.5 (100cycles) (Illumina). After sequencing, data was aligned to the human reference genome Hg38/GRCh38 using HISAT254 (v.2.1.0) and the number of reads per gene were calculated using HTSeq count55 (v.0.5.3). Gene expression differences, between the conditions, were determined using DESeq256 (v.1.22.2) with |log2(fold change)| > 1 and adjusted P value <0.05 cut-offs. Differentially expressed genes were ranked by log2(fold change expression) and used for gene set enrichment analysis (GSEA) on the Hallmark gene set from msigdbr (v.7.5.1), a previously published GR-activity signature16 or a newly developed PR-activity signature17 (Extended Data Table 3), using clusterProfiler57 (v.3.18.1), (pvalueCutoff = 0.05, pAdjustMethod = “BH”). GSEA enrichment plots have been generated using the plot.gsea function from the Rseb package48 (v.0.3.2).
Tumour RNA was extracted from FFPE tumour specimens from patients with breast cancer using the MasterPure Complete DNA and RNA Purification Kit (Lucigen, LGC Biosearch Technologies) following the manufacturer’s instructions. RNA quality was evaluated using Agilent RNA 6000 Nano Kit (Agilent Technologies) on the Agilent 2100 Bioanalyzer (Agilent Technologies). RNA-seq libraries were prepared using TruSeq Stranded Total RNA Library Prep Gold (20020598, Illumina) according to the manufacturer’s protocol and sequenced using 50 bp paired-end sequencing mode on Illumina Novaseq 6000 platform (Illumina). Differential gene expression analysis comparing Post- versus Pre- samples was performed using negative binomial distribution and Benjamini–Hochberg false discovery rate (FDR) with the Bioconductor package DESeq2, applying Wald tests on normalized counts to obtain log2(fold change) and P values for each gene. To evaluate the activity of pathways of interest (GR-activity signature and PR-activity signature16,17 (Extended Data Table 3) and Hallmark gene sets) we performed GSEA using the Bioconductor package clusterProfiler. GSEA was performed on genes ranked by the absolute value of log10(P value) scaled by the sign of log2(fold change), tested against gene lists of interest. The enrichment score and NES were computed for each gene set, and nominal P values were estimated by permutation testing. Multiple testing correction was applied using the Benjamini–Hochberg procedure to obtain FDR q values. To estimate the activity of pathways of interest at a single sample level we performed GSVA. Differences in enrichment scores between Post- and Pre- samples were determined by the paired Wilcoxon test. To correlate the activity of GR-activity signature and Hallmark gene sets of interest, their GSVA enrichment scores were correlated through Spearman’s linear correlation. All analyses were performed with R studio software (v.2023.03).
Proteomics
For protein digestion, frozen tissues were lysed in boiling guanidine HCl (GuHCl) lysis buffer as previously described58. Protein concentration was determined with a Pierce Coomassie (Bradford) Protein Assay Kit (Thermo Scientific), according to the manufacturer’s instructions. After dilution to 2 M GuHCl, aliquots corresponding to at least 1.05 mg of protein were digested twice (4 h and overnight) with trypsin (Sigma Aldrich) at 37 °C, enzyme:substrate ratio 1:75. Digestion was quenched by the addition of formic acid (final concentration 5%), after which the peptides were desalted on a Sep-Pak C18 cartridge (Waters). From the eluates, aliquots were collected for proteome analysis, the remainder being reserved for phosphoproteome analysis (not included in this work). Samples were vacuum dried and stored at −80 °C until LC–MS/MS analysis.
Prior to mass spectrometry analysis, the peptides were reconstituted in 2% formic acid. Peptide mixtures were analysed by nano LC–MS/MS on an Orbitrap Exploris 480 Mass Spectrometer equipped with an EASY-NLC 1200 system (Thermo Scientific). Samples were directly loaded onto the analytical column (ReproSil-Pur 120 C18-AQ, 2.4 μm, 75 μm × 500 mm, packed in house). Solvent A was 0.1% formic acid/water and solvent B was 0.1% formic acid/80% acetonitrile. Samples were eluted from the analytical column at a constant flow of 250 nl min−1. For single-run proteome a 90-min gradient was employed containing a 78-min linear increase from 6 to 30% solvent B, followed by a 10-min wash.
Raw data were analysed by DIA-NN (v.1.8)59 without a spectral library and with ‘Deep learning’ option enabled. The Swissprot Human database (20,395 entries, release 2021_04) was added for the library-free search. The Quantification strategy was set to Robust LC (high accuracy) and MBR option was enabled. The other settings were kept at the default values. The protein groups report from DIA-NN was used for downstream analysis in Perseus (v.1.6.15.0)60. Values were log2-transformed, after which proteins were filtered for at least 100% valid values in at least one sample group. Missing values were replaced by imputation based on a normal distribution using a width of 0.3 and a minimal downshift of 2.4. Differentially expressed proteins were determined using a Student’s t-test (threshold: FDR: 5% and S0: 0.1).
Differentially protein levels were ranked by log2(fold change) × P value and used for GSEA analysis on Hallmarks from the community-contributed functional database from the web-based Gene Set Analysis Toolkit (WebGestalt61). GSEA enrichment plots were generated as previously described.
ELISA
Mouse whole blood was collected in Eppendorf tubes. It was allowed to coagulate for 2 h at room temperature, centrifuged for 20 min at 4,000 rpm, then aliquoted into PCR tubes and stored at −80 °C until subsequent use. Whole blood from patients was collected in Vacuette Serum Clot Tubes (BD), centrifuged 20 min at 2,100 rpm then aliquoted into small tubes and stored at −80 °C until use. ELISA assays to detect mouse serum levels of cortisone and progesterone were purchased from R&D System and ALPCO respectively while ELISA assays to detect human serum levels of cortisol and progesterone were purchased from R&D System and Enzo, respectively.
Flow cytometry
Breast cancer nodules were macrodissected from TSAE1-bearing mice and processed to generate single-cell suspensions using the Tumour Dissociation Kit (Miltenyi Biotec, 130-096-730) in conjunction with the gentleMACS Octo Dissociator, following the manufacturers’ protocols. The resulting cell suspensions were passed through a 100-µm cell strainer and subsequently washed with fluorescence-activated cell sorting buffer (PBS containing 5% FBS). To ensure the removal of red blood cells, samples were treated with an erythrocyte lysis solution. Following this, samples were pre-incubated with an anti-CD16/CD32 antibody (1:400, 553142, BD Bioscience) and then stained with antibodies specific for extracellular markers, adhering to standard staining protocols. After staining for surface markers, cells were labelled with a live/dead viability dye and subsequently fixed and permeabilized using a fixation/permeabilization solution (eBioscience, Invitrogen) for intracellular staining. All antibodies utilized for flow cytometry were titrated to account for lot-dependent variations, as described in Extended Data Fig. 7f. Immune cell populations were classified as follows: lymphoid cells (CD45+CD11b−), myeloid cells (CD45+CD11b+), neutrophils (CD45+CD11b+Ly6G+), non-classical monocytes (CD45+CD11b+Ly6G−Ly6C−) and dendritic cells (CD45+F4/80−CD11chiMHCIIhi) (for gating strategy used, see Supplementary Fig. 1). Sample acquisition was performed using a five-laser Aurora spectral flow cytometer (Cytek Biosciences), and data analysis was conducted using FlowJo v.10 software.
Statistical analysis
Statistical analyses were performed using GraphPad Prism software v.10.4.1 (GraphPad Software) or in R v.4.0.2 (R Core Team 2020, https://www.R-project.org). Paired t-test or one-way analysis of variance (ANOVA) was used to calculate changes in the majority of the analyses (unless otherwise stated) and only P values <0.05 were considered significant. Two-tailed Wilcoxon signed rank test was used to compare cortisol plasmatic concentration measured before FMD (Pre-FMD) and after FMD (Post-FMD). For the animal experiments, a linear mixed-effects model was utilized to assess whether the mass volume exhibits a statistically significant trend relative to the treatment over time. The model was constructed using the two-way ANOVA or mixed-effect model (in case of missing data) from GraphPad Prism software v.10.4.1. The fixed-effects model matrix was generated by the interaction of time (measured in days after injection) and treatment. The random-effects term was specified as the interaction between the treatment group and the blocking factor of the mouse ID. Subsequent pairwise post hoc multiple comparisons were conducted using the same software. Statistical significance was determined by P values less than 0.05. Linear mixed-effect model results are presented in the Supplementary Information.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.