Mice
Female Thy1.2+ C57BL/6 (C57BL/6J) mice (aged 5–6 weeks) and female and male NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NSG) mice (aged 5–7 weeks) were purchased from Jackson Laboratory. TCR-transgenic Thy1.1+ Pmel-1 mice (B6.Cg-Thy1a/Cy Tg(TcraTcrb)8Rest/J) were originally purchased from the Jackson Laboratory and maintained in the Stanford University-Lorry Lokey (SIM1) facility. Mice were housed in animal facilities approved by the Association for the Assessment and Accreditation of Laboratory Care, under a 12 h–12 h dark–light cycle, with ambient temperature maintained at 22 ± 2 °C and humidity at 40–60%. Experimental procedures in mouse studies were approved by the Institutional Animal Care and Use Committee (IACUC) at the Stanford University (animal protocol ID 32279) and performed in accordance with the guidelines from the animal facility of Stanford University.
Cells and tumour models
The B16F10 mouse melanoma cell line, HEK293T cells, Raji cells, RAW 264.7 cells and J774A.1 cells were originally acquired from the American Type Culture Collection (ATCC). Platinum-E (Plat-E) and Platinum-GP (Plat-GP) retroviral Packaging Cell Line were purchased from Cell Biolabs. A375 and nRFP-M407 human melanoma cells were provided by A. Ribas. Mouse B cell lymphoma A20 cells were provided by R. S. Negrin and originally obtained from the ATCC. The NALM6 cell line was obtained from the ATCC. NALM6 cells were modified to express firefly luciferase (NALM6-Luc). Cell line authentication was performed by the supplier. All cell lines were confirmed to be mycoplasma free before use. B16F10, A375, HEK293T and Plat-E retroviral packaging cells were cultured in complete Dulbecco’s modified Eagle’s medium (DMEM) medium (Gibco/Thermo Fisher Scientific) supplemented with fetal bovine serum (FBS) (10% (v/v), Gibco/Thermo Fisher Scientific), penicillin–streptomycin (1% (v/v), Gibco/Thermo Fisher Scientific), l-glutamine (2 mM, Gibco/Thermo Fisher Scientific). Raji, nRFP-M407 and NALM6-Luc cells were maintained in RPMI-1640 supplemented with FBS (10% (v/v), Gibco/Thermo Fisher Scientific), and penicillin–streptomycin (1% (v/v), Gibco/Thermo Fisher Scientific). B16F10 tumour cells (3 × 105) were implanted s.c. into the right flanks of C57BL/6 WT mice to establish the syngeneic s.c. tumour models. Raji tumour cells (1 × 106) and A375 tumour cells (5 × 105) were inoculated s.c. into the right flanks of NSG mice to establish the xenograft tumour models. NALM6-Luc tumour cells (1 × 106) were injected i.v. into NSG mice to establish leukaemia xenograft models.
Mammalian expression vectors
cDNA encoding mouse orthogonal IL-2Rβ and geneblock cDNA encoding mouse ICDs of IL-2R, IL-4R, IL-7R, IL-9R, IL-21R, IFNAG2, IFNGR1, IFNLR1, IL-10R, IL-20R, IL-22R, EPOR and GCSFR (IDT) were cloned into the retroviral vector pMSCV-MCS-IRES-YFP by PCR and isothermal assembly (ITA). Geneblock cDNA encoding mouse NFIC, MAF and MAFB (IDT) were cloned into the retroviral vector pMSCV-MCS-IRES-YFP by ITA. Geneblock cDNA encoding mouse chimeric orthogonal IL-2Rβ-ECD–GCSFR (IDT) were cloned into the retroviral vector pMSCV-truncated EGFR(tEGFR) by ITA. Similarly, human orthogonal IL-2Rβ (ho2R), chimeric orthogonal IL-2Rβ-ECD–IL-4R-ICD (ho4R), chimeric orthogonal IL-2Rβ-ECD–IL-22R-ICD (ho22R) and chimeric orthogonal IL-2Rβ-ECD–GCSFR-ICD (hoGCSFR) constructs were inserted into the pMSCV vector using the same methods. The STAT3-binding motif (YRHQ) and C-terminal domain (CTD) of IL-22R were fused at the C terminus of the mouse CD19 CAR construct (1D3) by ITA. The human CD19 CAR construct (FMC63) was acquired from Addgene (127889). The constructs for TCRs targeting NY-ESO-1 (1G4) in the pMSGV-tEGFR vector were provided by A. Ribas.
Retrovirus production
HEK293T cells were seeded at 3 × 106 cells per 10 cm tissue culture dish. After incubation overnight, the medium was replaced with prewarmed DMEM with FBS (5%, v/v), penicillin–streptomycin (0.5%, v/v). For each transfection, 8.3 µg of plasmid (5 µg of plasmid containing the gene of interest plus 3.3 µg of pCL-Eco packaging plasmid) was added to 830 µl of Opti-MEM I reduced rerum medium (Gibco/Thermo Fisher Scientific), followed by the gradual addition of 25 µl of TurboFect transfection reagent (Thermo Fisher Scientific) with gentle vortexing. After incubation for 20 min, the transfection mixture was gently added to HEK293T cells. The culture medium was replaced 16 h later and, after an additional 24 h, the medium was collected, clarified by centrifugation and passed through a 0.45-μm filter. If not used immediately, virus was frozen at −80 °C for later use. To generate retrovirus for the transduction of human T cells, Plat-GP packaging cells (Cell Biolabs) were transfected with relevant plasmids using FuGENE HD transfection reagent (Promega). For each transfection, 20 µg of plasmid (consisting of 15 µg of expression plasmid plus 5 µg of pLTR-RD114A envelope plasmid) was added to 1 ml of Opti-MEM I medium, followed by addition of 60 µl of FuGENE HD transfection reagent. The culture medium was replaced 24 h later and, after an additional 24 h, the medium was collected, clarified by centrifugation and passed through a 0.45-μm filter before storage at −80 °C.
Activation and transduction of primary mouse T cells
Spleens from C57BL/6 WT, pmel-1 mice were disintegrated mechanically and filtered through a 70-μm strainer (Thermo Fisher Scientific). Red blood cells (RBCs) were lysed with ACK lysis buffer (2 ml per spleen, Gibco/Thermo Fisher Scientific) for 5 min at 25 °C. The splenocytes were washed once with T cell medium, which contained RPMI-1640 (Gibco), FBS (10% (v/v), Gibco/Thermo Fisher Scientific), HEPES (25 mM, Gibco/Thermo Fisher Scientific), penicillin–streptomycin (1% (v/v), Gibco/Thermo Fisher Scientific), sodium pyruvate (1% (v/v), Gibco/Thermo Fisher Scientific), MEM non-essential amino acids solution (1% (v/v), Gibco/Thermo Fisher Scientific) and 2-mercaptoethanol (0.1% (v/v), Gibco/Thermo Fisher Scientific), and were then resuspended at a cell density of 2 × 106 cells per ml in a complete RPMI-1640 medium supplemented with MSA–mIL-2 (100 U ml−1). For WT T cell activation, cells were activated with plate-bound anti-mouse CD3ε (2.5 μg ml−1, 145-2C11, BioLegend) and soluble anti-mouse CD28 (5 μg ml−1, 37.51, BioXCell) for 24 h. Pmel-1 T cells were activated with hgp10025–33 peptide (1 μM, GenScript). Cells were cultured at 37 °C for 24 h. T cells were enriched by using Ficoll-Paque PLUS (Cytiva) and seeded onto precoated six-well plates at 3 × 106 cells per well in complete RPMI-1640 medium (3 ml per well). Then, 1 day before transduction, 12-well tissue culture plates were coated with retronectin (25 μg ml−1, Takara) and placed into a 4 °C refrigerator overnight. The next day, the plates were blocked with 0.5% FBS in PBS for 30 min and washed with PBS. Activated T cells (2 × 106 in 1 ml T cell medium) and viral supernatant (3 ml) were added to each well, along with MSA–mIL-2 (100 U ml−1), and centrifuged at 2,500 rpm for 1.5 h at 32 °C. After incubating overnight, T cells were collected and expanded in a T cell medium supplemented with MSA–mIL-2 (100 U ml−1). On day 3, the transduction efficiency was assessed on the basis of the expression of YFP using flow cytometry. Untransduced (NT) T cells activated and cultured in parallel were used as a control. Cells were collected for in vitro assays or i.v. injection 2 days after spinfection.
Activation and transduction of primary human T cells
Primary human peripheral blood mononuclear cells isolated from a healthy human donor by leukapheresis were thawed and resuspended at a cell density of 2 × 106 cells per ml in T cell medium supplemented with human IL-2 (10 ng ml−1, PeproTech). For human T cell activation, cells were activated with plate-bound anti-human CD3ε (1 μg ml−1, OKT-3, BioXCell) and soluble anti-human CD28 (5 μg ml−1, 9.3, BioXCell) for 48 h. Activated T cells were collected and transduced on 12-well plates coated with retronectin (25 μg ml−1, Takara) and loaded with 1.5 ml per well of each retrovirus (encoding NY-ESO-1 TCR clone 1G4 and ho2R, ho22R or hoGCSFR, or CD19 CAR clone FMC63 and ho22R or hoGCSFR) by spinfection. After incubating overnight, T cells were collected and expanded in a T cell medium supplemented with human IL-2 (10 ng ml−1). Cells were stained with anti-human EGFR antibody (BioLegend, recognizes tEGFR in the NY-ESO-1 TCR construct) or anti-MYC tag antibody (Cell Signaling, recognizes the MYC-tag in the CD19 CAR construct). The transduction efficiency was detected on the basis of the expression of YFP, EGFR and MYC. NT T cells activated and cultured in parallel were used as control. Cells were expanded and collected for in vitro assays or i.v. injection 10–12 days after activation.
CRISPR–Cas9 gene editing
Activated ho22R T cells were transduced with lentiCRISPR vectors encoding sgRNAs targeting BACH2 (5′-CATCCTTCCGGCACACAAAC-3′, 5′-CTAGCAACAGCCTCAAGCCG-3′, 5′-ACGTGACTTTGATCGTGGAG-3′) and expressing mCherry as a reporter. Transduction was performed by spinfection as described above. mCherry+ cells were sorted 72 h later for flow cytometry analysis and quantitative PCR. For ho4R T cells, sgRNAs targeting GATA3 (5′-CCTACTACGGAAACTCGGTC-3′, 5′-GGAGCTGTACTCGGGCACGT-3′, 5′-TCGACGAGGAGGCTCCACCC-3′) were complexed with Alt-R Cas9 protein to form ribonucleoproteins, followed by electroporation using the Lonza 4D-Nucleofector system. Cells were immediately transferred to IL-2 supplemented T cell medium, and the KO efficiency was assessed using flow cytometry.
Flow cytometry analyses
For surface marker staining, cells were collected into U-bottom 96-well plates (Thermo Fisher Scientific), blocked with anti-mouse CD16/32 antibody (BioLegend) or human TruStain FcX (BioLegend), and incubated with the indicated antibodies at 4 °C for 20 min, followed by live/dead staining by 4′,6-diamidino-2-phenylindole (DAPI, Thermo Fisher Scientific). Cells were then washed and resuspended with FACS buffer (PBS containing 0.2% BSA, Sigma-Aldrich) for flow cytometry analyses. For pSTAT staining, primary mouse or human T cells were rested in T cell medium lacking IL-2 for 24 h before signalling assays. Cells were plated in a 96-well round-bottom plate in 50 μl T cell medium. Cells were stimulated by addition of MSA fused to mouse orthogonal IL-2 (MSA–oIL-2) or MSA fused to human orthogonal IL-2 (MSA–hoIL-2) for 15 min at 37 °C, and the reaction was terminated by fixation with 1.5% paraformaldehyde for 15 min at room temperature with agitation. Cells were washed and permeabilized with ice-cold 100% methanol for 60 min on ice. Next, cells were washed with FACS buffer before staining with pSTAT antibodies for 1 h at 4 °C in the dark. Cells were washed and resuspended in FACS buffer for flow cytometry analyses. For intracellular cytokine staining, cells were first stimulated by a cell stimulation cocktail (protein transport inhibitors included, Invitrogen/Thermo Fisher Scientific) at 37 °C for 5 h. After stimulation, cells were first stained for surface markers and Zombie Violet Fixable Dye (BioLegend), then fixed and permeabilized using the Cytofix/Cytoperm Fixation/Permeabilization Solution Kit (BD Biosciences). Intracellular staining with the indicated antibodies was performed according to the manufacturer’s protocol. For TF or BrdU staining, cells were first stained for surface markers and Zombie Violet Fixable Dye. Next, cells were fixed and permeabilized using the FOXP3/Transcription Factor Staining Buffer Set (eBioscience) for TFs, or the BD Pharmingen APC BrDU Kit (BD Biosciences) for BrdU according to the manufacturer’s instructions, followed by incubation with the indicated antibodies for intracellular staining. Cells were detected using the CytoFlex (Beckman Coulter) system. Analyses were performed using FlowJo (v.10.10.0).
Antibodies and reagents for flow cytometry
The following antibodies or staining reagents were purchased from BioLegend: mouse CD16/32 (93, 101302), mouse CD90.1/Thy-1.1 (OX-7, 202533), mouse CD8β (YTS156.7.7, 126606), mouse CD8β (YTS156.7.7, 126614), mouse CD45.2 (104, 109808), mouse Ki-67 (16A8, 652413), mouse SCA1 (D7,108142), mouse CD44 (IM7, 103030), mouse CD62L (MEL-14, 104428), mouse PD-1 (29F.1A12, 135220), mouse IFNγ (XMG1.2, 505850), mouse TNF (MP6-XT22, 506329), mouse/human GZMB (QA16A02, 372214), mouse/human KLRG1 (2F1/KLRG1, 138419), mouse IL-7Rα (A7R34, 135022), mouse CD122 (TM-β1, 123210), mouse SLAMF6 (330-AJ, 134606), mouse Mac-1 (M1/70, 101228), mouse CD14 (Sa14-2, 123335), mouse CD64 (X54-5/7.1, 139303), mouse Gr-1 (1A8, 127648), mouse SIRPα (P84, 144011), human EGFR (AY13, 352906), human CD3 (HIT3a, 300308), human CD4 (SK3, 344646), human CD8 (SK1, 344724), human IL-13 (JES10-5A2, 501914), human IL-4 (MP4-25D2, 500845), human IL-5 (TRFK5, 504304), mouse/human GATA3 (W19195B, 386906), human CXCR3 (G025H7, 353737), human CCR4 (L291H4, 359443), human CD62L (DREG-56, 304830), human CD95 (DX2, 305622), human CD45RA (HI100, 304120), human CD27 (O323, 302832), human CCR7 (G043H7, 353214), human CD45RO (UCHL1, 304228), human CD66b (G10F5, 305121), human LAG3 (11C3C65, 369322), human IFNγ (4S.B3, 502530), human Ki-67 (Ki-67, 350526), human CD39 (A1, 328240), human TIM3 (F38-2E2, 345026), human TruStain FcX (422302) and Zombie Violet Fixable Viability Kit (423114). The following antibodies or staining reagents were purchased from BD Biosciences: pSTAT3 (4/pSTAT3, 557815), pSTAT4 (38/pSTAT4, 558137), pSTAT5 (47/STAT5, 612599), mouse pSTAT6 (J71-773.58.11, 558252), human pSTAT6 (23/STAT6, 612701) and the BD Pharmingen APC BrdU Kit (552598). The following antibodies were purchased from Cell Signaling: pSTAT1 (58D6, 8009S) and MYC-tag (9B11, 3739/2233S). DAPI was purchased from Thermo Fisher Scientific. For flow cytometry staining, surface marker antibodies were used at a 1:200 dilution, intracellular antibodies at 1:100 and pSTAT antibodies at 1:50.
Protein production
The cDNAs encoding mouse and human orthogonal IL-2 was cloned into the mammalian expression vector pD649, which includes a C-terminal 8×His tag for affinity purification. DNA encoding MSA was purchased from Integrated DNA Technologies (IDT) and cloned into pD649 as an N-terminal fusion. Mammalian expression DNA constructs were transfected into HEK293F cells using the Expi293 Expression System (BD Biosciences) for secretion and purified from the clarified supernatant by nickel affinity resin (Ni-NTA, Qiagen) followed by size-exclusion chromatography with a Superdex-200 column (Cytiva) and formulated in sterile PBS for injection. Endotoxin was removed using the Proteus NoEndo HC Spin column kit according to the manufacturer’s recommendations (VivaProducts) and endotoxin removal was confirmed using the Pierce LAL Chromogenic Endotoxin Quantification Kit (Thermo Fisher Scientific). Proteins were concentrated, flash-frozen in liquid nitrogen and stored at −80 °C until ready for use.
In vivo anti-tumour therapy studies
C57BL/6 WT mice bearing established s.c. B16F10 tumours were sublethally lymphodepleted by total body irradiation (5 Gy) on day 5. On day 6, mice received i.v. adoptive transfer of 3 × 106 pmel-1 T cells (either NT or transduced with o2R, o4R, o7R, o9R, o21R, o10R, o20R, o22R, oIFNAR2, oIFNGR1, oIFNLR1, oEPOR or oGCSFR) followed by i.p. administration of MSA–oIL-2 (2.5 × 104 U per day) or PBS control every other day until day 20. For the s.c. Raji tumour model, NSG mice received i.v. adoptive transfer of 1 × 106 CD19 CAR T cells, ho22R CD19 CAR T cells or hoGCSFR CD19 CAR T cells on day 7 after tumour inoculation followed by i.p. administration of MSA–hoIL-2 (2.5 × 104 U per day) or PBS control every other day until day 29. For the s.c. A375 tumour model, NSG mice received i.v. adoptive transfer of 3 × 106 NY-ESO-1 TCR-T cells (WT NY-ESO-1 TCR-T cells, or ho2R-, ho4R-, ho22R- or hoGCSFR-expressing NY-ESO-1 TCR-T cells) on day 6 after tumour inoculation followed by i.p. administration of MSA–hoIL-2 (1 × 105 U per day) or PBS control every other day until day 20. For the s.c. tumour model, the tumour area was calculated using the formula area = length × width from calliper measurements of two orthogonal diameters. For the NALM6-Luc leukaemia model, NSG mice received i.v. adoptive transfer of 4 × 105 CD19 CAR T cells, ho22R CD19 CAR T cells or hoGCSFR CD19 CAR T cells on day 5 after tumour inoculation followed by i.p. administration of MSA–hoIL-2 (2.5 × 104 U per day) or PBS control every day until day 18. Mice were anaesthetized and i.p. injected with bioluminescent substrate d-luciferin potassium salt (30 μg ml−1, 100 µl, GoldBio) prediluted in PBS. Then, 10 min after injection, the mice were subjected to luminescence imaging using the Xenogen IVIS fluorescence/bioluminescence imaging system for tumour growth monitoring. Sample size was not predetermined statistically but was based on standards commonly used in previous studies. Mice were assigned to experimental groups to ensure uniform starting tumour sizes. No blinding was performed during the study. Post-therapy survival of mice was monitored for at least 90 days after tumour inoculation. Mice were euthanized when body weight loss was beyond 15% of the baseline weight, the tumour area reached 200 mm2 or any signs of discomfort were detected by the investigators or as recommended by the caretaker who monitored the mice every day.
Immunophenotyping by flow cytometry
C57BL/6 mice were inoculated s.c. with B16F10 tumour cells (1 × 106), sublethally lymphodepleted by irradiation on day 7 and received i.v. adoptive transfer of 3 × 106 pmel-1 T cells (either NT or transduced with o2R, o4R, o7R, o9R, o21R, o10R, o20R, o22R, oIFNAR2, oIFNGR1, oIFNLR1, oEPOR or oGCSFR) on day 8 after tumour inoculation followed by i.p. administration of MSA–oIL-2 (2.5 × 104 U per day) or PBS control every other day until day 16. Mice received i.p. administration of bromodeoxyuridine (BrdU) (1 mg per 100 µl, BD Pharmingen) on day 16. On day 17, mice were euthanized, and tumours, TDLNs and spleens were collected. NSG mice were inoculated s.c. with A375 cells (1 × 106) and received i.v. ACT of 3 × 106 NY-ESO-1 TCR-T cells (WT NY-ESO-1 TCR-T cells, or ho2R-, ho22R- or hoGCSFR-expressing NY-ESO-1 TCR-T cells) on day 10 after tumour inoculation followed by i.p. administration of MSA–hoIL-2 (2.5 × 104 U, 3 doses, 1 × 105 U, 3 doses) or PBS control every other day until day 20. On day 21, mice were euthanized, tumours and spleens were collected. For the Raji tumour model, NSG mice were inoculated (s.c.) with Raji cells (2 × 106) and received an i.v. adoptive transfer of 1 × 106 NT, ho22R or hoGCSFR CD19 CAR T cells on day 11, followed by i.p. administration of MSA–hoIL-2 (2.5 × 104 U) or PBS every other day until day 25. On day 26, the mice were euthanized, and tumours were collected for flow cytometry analysis. Collected tumours were weighed, mechanically minced and digested in RPMI-1640 medium supplemented with collagenase type IV (1 mg ml−1, Gibco/Thermo Fisher Scientific), dispase II (100 μg ml−1, Sigma-Aldrich), hyalurondase (100 μg ml−1, Sigma-Aldrich) and DNase I (100 μg ml−1, Sigma-Aldrich) at 37 °C for 60 min. RBC lysis was performed on the digested tumour samples with ACK lysing buffer. Tumour-infiltrating leukocytes were then enriched by Percoll (Cytiva) density-gradient centrifugation, resuspended in PBS with BSA (0.2%, w/v), stained with the indicated antibodies and analysed by flow cytometry. Spleens were ground and filtered through a 70-μm strainer (Thermo Fisher Scientific). RBC lysis was performed on the spleen samples with ACK lysing buffer (2 ml per spleen, Gibco/Thermo Fisher Scientific) and then resuspended in PBS with BSA (0.2%, w/v). TDLNs were ground and filtered through a 70-μm strainer (Thermo Fisher Scientific) and then resuspended in PBS with BSA (0.2%, w/v). Cells collected from blood, spleen, tumour and TDLNs were stained with the indicated antibodies and analysed on the CytoFLEX flow cytometer (Beckman Coulter).
Phagocytosis assay
oGCSFR-transduced pmel-1 CD8+ T cells were maintained in MSA–oIL-2 (500 nM) for 72 h. Live CD8+Mac-1− and CD8+Mac-1+ cells were sorted using the Aria II sorter (BD Biosciences) at the Stanford Shared FACS Facility, with NT cells sorted as controls. Phagocytic activity was assessed using the pHrodo Red S. aureus BioParticles Conjugate (Thermo Fisher Scientific) according to the manufacturer’s protocol. In brief, sorted cells and J774A.1 cells were seeded into a 96-well U-bottom plate and incubated with a suspension of pHrodo Red S. aureus BioParticles for 30 min. After incubation, extracellular fluorescent probes were washed off, and the cells were stained for surface markers and with DAPI. Fluorescence was measured on the CytoFLEX flow cytometer (Beckman Coulter). For real-time phagocytosis analysis, sorted cells were seeded into a 96-well flat-bottom plate (10,000 cells per well) and incubated with pHrodo Red S. aureus BioParticles (Thermo Fisher Scientific). Fluorescence images of each well were captured every 20 min using the IncuCyte live imaging system (Essen Bioscience), and phagocytosis was quantified as the percentage confluence. For the live bacterial phagocytosis assay, NT or oGCSFR-tEGFR-transduced CD8+ T cells were seeded into a 96-well flat-bottom plate (200,000 cells per well) and incubated with GFP+ L. monocytogenes (1/2a) derived from FDA LS808 (Microbiologics) at a series of concentrations, with 10× dilution for 2 h. Next, the supernatant was removed, and the cells were cultured in gentamicin-containing RPMI complete medium at 37 °C for an additional 22 h. GFP signals from each well were analysed on the CytoFLEX flow cytometer (Beckman Coulter). Live-cell imaging without sorting was performed using the Leica TCS SP8 Confocal system with a white-light laser. Fixed cells were sorted and visualized using the Zeiss Elyra7 lattice SIM microscope.
ADCP assay
A20 cells were labelled with FarRed using the Invitrogen CellTrace Far Red Cell Proliferation Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Labelled A20 cells (5 × 106) were then incubated with InVivoMAb anti-mouse CD19 (5 μg ml−1, ID3, BioXCell) in 1.25 ml RPMI medium at 37 °C for 10 min. No-ICD o2R and oGCSFR-transduced pmel-1 CD8+ T cells were maintained in MSA–oIL-2 (500 nM) for 72 h. For the blocking experiment, T cells were pretreated with a blocking cocktail containing anti-mouse CD64 (BioLegend, X54-5/7.1) and anti-mouse CD16/32 (BioLegend, 93) at 1 μg ml−1. RAW264.7 cells, NT pmel-1 CD8+ T cells, no-ICD o2R-transduced pmel-1 CD8+ T cells and oGCSFR-transduced pmel-1 CD8+ T cells were seeded into a 96-well round-bottom plate at 5 × 104 cells per well. FarRed+ A20 target cells were added at a target:effector ratio of 2:1, and co-cultures were incubated for 2 h. After incubation, cells were stained for surface markers, then with DAPI (Sigma-Aldrich) and then washed with FACS buffer and analysed using the CytoFLEX flow cytometer (Beckman Coulter). Confocal imaging (Leica TCS SP8 with a white-light laser) was performed at a effector:target (E:T) ratio of 1:1, and co-cultures were incubated for 24 h.
Human T cell differentiation assay
Human T cells were activated on day 0 using ImmunoCult Human CD3/CD28 T Cell Activator (StemCell) and retrovirally transduced to generate NY-ESO-1 TCR-T cells or ho4R NY-ESO-1 TCR-T cells on day 2. After transduction, ho4R NY-ESO-1 TCR-T cells or NY-ESO-1 TCR-T cells were cultured in RPMI complete medium supplemented with MSA–hoIL-2 (100 nM) or recombinant human IL-2 (10 nM, Peprotech), respectively. The cell density was adjusted to 1 × 106 cells per ml every 2 days as needed with fresh complete medium supplemented with MSA–hoIL-2 or recombinant human IL-2. After 7 days of incubation, cells were reactivated using ImmunoCult Human CD3/CD28 T Cell Activator and cultured for an additional 7 days with cell density adjustments performed as previously described. On day 14, differentiated cells were stained for the indicated markers and analysed using the CytoFlex flow cytometer (Beckman Coulter).
Cell killing assays
CD19 CAR T cells (either NT or transduced with ho22R or hoGCSFR) were co-cultured with target cells (NAML6-Luc or Raji) at the different E:T ratios in the presence or absence of MSA–hoIL-2 (100 nM). After co-culture for 24 h, the cells in the plates were collected and analysed by flow cytometry to determine the viability of tumour cells. NT T cells, ho4R NY-ESO-1 TCR-T cells, ho4R NY-ESO-1 TCR-T cells (GATA3 KO) or ho4R NY-ESO-1 TCR-T cells with anti-IL-4 (BioXCell, MP4-25D2, 2 μg ml−1), anti-IL-5 (BioXCell, TRFK5, 2 μg ml−1), anti-IL-13 (BioXCell, tralokinumab, 2 μg ml−1) or their combination (2 μg ml−1 each) were co-cultured with A375 cells at an E:T ratio of 1:2 for 48 h. Viable tumour cell counts were assessed by flow cytometry. Human melanoma cells (nRFP-M407, 1 × 106 per well) were plated in six-well plates. NT T cells, NY-ESO-1 TCR-T cells or NY-ESO-1 TCR-T cells co-transduced with ho2R, ho22R or hoGCSFR were added in duplicate at a 1:1 E:T ratio with MSA–hoIL-2 (100 nM). Every 48 h, the cells were collected, washed, resuspended in fresh MSA–hoIL-2 and added to the nRFP-M407 tumour cells at a 1:1 ratio. For real-time cell killing analysis, after each 48 h co-culture, the T cells were collected from the six-well plates, resuspended in fresh MSA–hoIL-2 (100 nM) and added to nRFP-M407-preseeded flat-bottom 96-well plates (40,000 cells per well) at a 1:1 E:T ratio and fluorescence images were obtained for each well every 3 h using the IncuCyte live imaging system (Essen Bioscience) and quantified by percentage confluence.
Human T cell repetitive stimulation assays
Human melanoma cells (A375, 1 × 106 per well) were plated in six-well plates. NT T cells, NY-ESO-1 TCR-T cells or NY-ESO-1 TCR-T cells co-transduced with either ho22R or hoGCSFR were added in duplicate at a 1:1 E:T ratio with MSA–hoIL-2 (100 nM). Every 48 h, the cells were collected, washed, resuspended in fresh MSA–hoIL-2 and were rechallenged with A375 tumour cells (1 × 106). After the three rounds of stimulation with target cells, the T cells were collected for phenotyping by flow cytometry.
RNA-seq sample preparation and data analysis
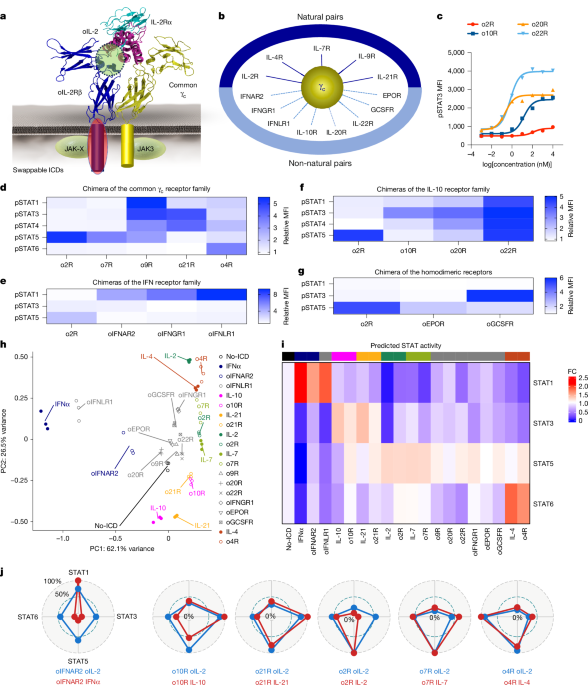
WT mouse T cells transduced with orthogonal receptors were sorted using the Aria II sorter (BD Biosciences) at the Stanford Shared FACS Facility. No-ICD o2R-transduced cells were sorted as controls. After sorting, cells were recovered in MSA–mIL-2 (100 U ml−1) for 24 h, subjected to IL-2 starvation for another 24 h and then stimulated with 5 μM MSA–oIL-2 or 10 nM recombinant cytokines (IL-2, IL-4, IL-7, IL-21, IL-10 and IFNα; Peprotech) for 6 h. Total RNA was extracted using the Quick-RNA 96 Kit (Zymo Research). Libraries were synthesized using the mRNA library preparation (poly A enrichment) kit according to the manufacturer’s instructions. Libraries were pooled and sequenced on the NovaSeq X Plus Series (PE150). Reads were aligned to the mouse reference genome (mm10) using Rsubread (v.2.18.0)58. Gene expression was quantified with featureCounts. We first conducted differential expression analysis between the no-ICD o2R plus MSA–oIL-2 condition and cytokine-treated conditions (IFNα, IL-10, IL-21, IL-2, IL-7 and IL-4) to define differentially regulated genes induced or repressed by these canonical cytokines. DESeq2 (v.1.48.1) was used for this analysis, and genes with FDR-adjusted P values < 0.05 were considered to be differentially regulated. IFNα- and IL-4-driven genes were interpreted as STAT1- and STAT6-driven genes. For STAT3-driven genes, intersection was taken from IL-10- and IL-21-driven genes. For STAT5-driven genes, intersection was taken from IL-2- and IL-7-driven genes. We next computed the transcripts per million (TPM) for all genes across all samples. We then computed Pearson’s correlation coefficients to each of the reference samples (for example, samples treated with IL-10 and IL-21 in the case of STAT3) by using only TPM values of genes driven by the STAT of interest (for example, STAT3) but not driven by other STATs (such as STAT1, STAT5 or STAT6). As there are multiple reference samples, we took the mean of those correlation coefficients for each individual sample. We considered these averaged correlation coefficients to be STAT scores. PCA was performed with STAT scores. Heat-map and radar charts were generated using the fold-change values of these STAT scores compared with the no-ICD o2R plus MSA–oIL-2 condition.
scRNA-seq sample preparation
C57BL/6 mice were inoculated s.c. with B16F10 tumour cells (1 × 106), sublethally lymphodepleted by irradiation on day 7 and received i.v. adoptive transfer of 3 × 106 pmel-1 T cells (NT or transduced with o2R, o4R, o20R, o22R or oGCSFR) on day 8 after tumour inoculation followed by i.p. administration of MSA–oIL-2 (2.5 × 104 U per day) or PBS control every other day until day 16. On day 17, the mice were euthanized, tumours were minced and dissociated using a mouse tumour dissociation kit (Miltenyi Biotec) and the gentleMACS Octo Dissociator (Miltenyi Biotec). TILs were first enriched by density-gradient centrifugation against Percoll (Cytiva), and then stained for surface markers and with DAPI (Thermo Fisher Scientific). Thy1.1+CD8+ or Thy1.1+CD8+YFP+ T cells were sorted using the Aria II sorter (BD Biosciences) at the Stanford Share FACS Facility. Sorted cells were subjected to single-cell encapsulation using the Chromium Single Cell Instrument and reagents. A Chromium Next GEM Chip G was loaded with the appropriate number of cells, and the sequencing libraries were prepared using 10x Genomics reagents according to the manufacturer’s instructions and passed quality control. In brief, an emulsion encapsulating single cells into droplets with reagents and gel beads containing a unique molecular identifier, reverse transcription reagents and cell barcoding oligonucleotides was generated. cDNAs were obtained and amplified after droplets broke. For the 3′ Gene Expression library, the cDNA was fragmented, ligated to a sequencing adaptor and PCR amplified. The generated 3′ Gene Expression libraries were sequenced using the NovaSeq 6000 system with a sequencing depth of >20,000 paired-end reads per cell. The fastq files were generated by Cell Ranger (v.7.1.0) mkfastq from 10x Genomics, and primary data analysis using a custom reference package based on the mm10 reference genome.
Analysis of scRNA-seq data
The gene expression matrix was first processed using Seurat (v.5.1.0). For each dataset, we excluded cells containing fewer than 200 genes (to remove debris, empty droplets and low-quality cells) and also cells in which >20% of transcripts were derived from mitochondrial RNA, leaving 32,075 cells. All expression data were normalized by log transformation. Seurat was used to first normalize gene expression count data using the NormalizeData and ScaleData functions. Then, the FindVariableFeatures function was used to select the top 2,000 variable genes and PCA was performed. Cell clusters were identified using the FindNeighbors and FindClusters functions. After clustering, we omitted two clusters (n = 597 cells) with low Cd8 and Thy1 expression to exclude non pmel-1 T cell contaminants. We also omitted one cluster with low Ptprc and high Hbb expression and high ribosomal RNA content (n = 2,346 cells) to further exclude any non-leukocytes, and one cluster with negligible cell count (n = 124 cells). The final Seurat dataset contained 29,008 cells.
TF activity prediction was conducted using the pySCENIC (v.0.12.1) docker distribution with the default parameter settings. A total of 241 regulons was identified, with corresponding TFs annotated. The area under the receiver-operator curve (AUC) scores for all TFs were stored in a designated assay slot (termed scenic) and used for UMAP computation with the Seurat RunUMAP function with the default parameters. For unsupervised clustering, cells were first embedded into a ten-dimentional UMAP space, and clusters were then identified using the FindCluster function with the resolution parameter set to 0.02. For the search for marker TFs, the FindAllMarkers function was used with the min.pct parameter set to 0.50 (that is, at least 50% of cells have positive TF activity), and TFs with FDR-adjusted P value < 0.05 and average log2-transformed fold-change > 1 were considered to be markers. Up to ten TFs, sorted based on the average log2-transformed fold-change values, are shown in the main figure. For marker gene analysis, the FindAllMarkers function was used with min.pct set to 0.10 for the gene expression matrix. Standard Seurat functionalities were used for data visualization. Intercellular communication prediction was performed using LIANA (v.1.5.1)59. The built-in MouseConsensus ligand–receptor database was used. Five algorithms (natmi, connectome, logfc, sca and cellphonedb) were used, and only LR interactions concordant between all algorithms were retained.
ATAC-seq sample preparation
NSG mice were inoculated s.c. with A375 cells (1 × 106) and received i.v. ACT of 3 × 106 NY-ESO-1 TCR-T cells (WT NY-ESO-1 TCR-T cells, or ho22R- or hoGCSFR-expressing NY-ESO-1 TCR-T cells) on day 10 after tumour inoculation followed by i.p. administration of MSA–hoIL-2 (2.5 × 104 U, 3 doses, 1 × 105 U, 3 doses) or PBS control every other day until day 20. On day 21, the mice were euthanized and tumours were minced and dissociated using the human tumour dissociation kit (Miltenyi Biotec) and the gentleMACS Octo Dissociator (Miltenyi Biotec). TILs were first enriched by density-gradient centrifugation against Percoll (GE healthcare), and were then stained for surface markers and with DAPI (Sigma-Aldrich). CD3+EGFR+ or CD3+EGFR+YFP+ T cells were sorted using the Aria II sorter (BD Biosciences) at the Stanford Share FACS Facility. For ATAC-seq library preparation, intact nuclei from sorted cells were treated with a hyperactive Tn5 transposase mutant. This transposase simultaneously tags the target DNA with sequencing adapters and fragments the DNA. The tagmented DNA is then purified and amplified using indexed primers to generate libraries. Equal amounts of each sample were pooled and subjected to 50 bp paired-end sequencing on the NovaSeqX sequencer.
Chromatin accessibility analysis
After routine quality control with FastQC, reads were mapped onto the reference genome (hg19) using RSubread (v.2.18.0) in the DNA mode58. The aligned reads were then subjected to peak calling using MACS3 (v.3.0.1)60. Downstream analyses were performed in R (v.4.4). Peaks were annotated with their genomic locations and associated genes using ChIPseeker (v.1.44.0)61. A set of non-redundant peaks across all samples was defined using ChIPQC. The reads aligned with each region were then counted using the summarizeOverlaps function, and the count matrix was analysed with DESeq2 (v.1.48.1)62. Count data normalized by variance-stabilizing transformation were subjected to PCA for visualization. Differential chromatin accessibility analysis was conducted according to the standard DESeq2 workflow between each transduction group (biological triplicates). Differential TF motif enrichment analysis was performed using chromVAR (v.1.30.1)63. Here, all samples were analysed simultaneously, and the TF motifs most significantly variably regulated among transduction group were chosen for heat-map analysis. Cis-regulatory element enrichment analysis were performed with GREAT (v.2.10.0)64 for each sample separately. Gene Ontology terms significantly enriched in at least one sample in a given transduction group were considered. Terms enriched in the hoGCSFR group but not in the ho22R or the NT group were manually inspected.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 9 (GraphPad software), except bulk RNA-seq, scRNA-seq and ATAC-seq data, which were analysed with R (described above). All values and error bars are shown as mean ± s.e.m. Comparisons of two groups were performed by using two-tailed unpaired Student’s t-tests. Comparisons of multiple groups were performed using one-way ANOVA with Tukey’s multiple-comparison test unless otherwise indicated. Experiments that involved repeated measures over a time course, such as tumour growth, were performed using two-way ANOVA with Tukey’s multiple-comparison post-test. Survival data were analysed using the log-rank (Mantel–Cox) test. No statistically significant differences were considered when P values were larger than 0.05.
Statistics and reproducibility
Animal studies were independently repeated 2–3 times. For groups with more than five mice, the total number of animals was pooled from two independent experiments. In vitro experiments were independently repeated 2–4 times with consistent results.
Ethics statement
Experimental procedures in mouse studies were approved by the Institutional Animal Care and Use Committee (IACUC) at the Stanford University (animal protocol ID 32279) and performed in accordance with the guidelines from the animal facility of Stanford University. Primary T lymphocytes from healthy donors were provided by the Stanford Blood Center. Ethical approval pertaining to T cell donors was obtained by the Stanford Blood Center.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.