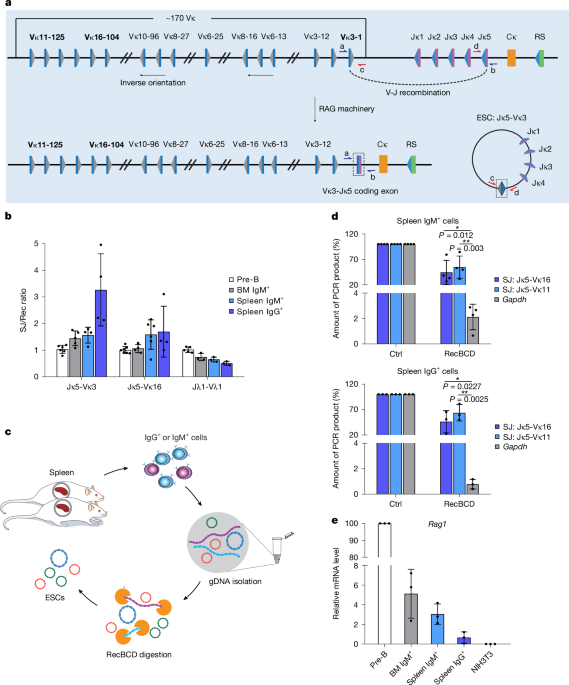
Purification of mouse B cells
Non-transgenic CBA/C57BL/6 J mice were obtained from the University of Leeds animal facility, which is a full barrier facility, with a light/dark cycle of 12 h on/12 h off, an ambient temperature of 21 °C (range 20–22 °C) and 45–65% humidity. No more than six animals are housed per cage and all mice are free of common pathogens, including murine norovirus, Pasteurella and Helicobacter. Animal procedures were performed under Home Office licence P3ED6C7F8, following review by the University of Leeds ethics committee. Mouse femurs and spleens were collected from 5- to 7-week-old mice, using 12 mice in total to minimize animal numbers used, according to 3Rs principles. Roughly equal numbers of male and female mice were used; randomization and blinding were not necessary, as all mice were wild type. Bone marrow cells were flushed from femurs with PBS whereas splenocytes were prepared by flushing cells from finely diced pieces of spleen with PBS through a 50-µm cell strainer. Following preparation of single-cell suspensions in PBS, bone marrow cells and splenocytes were centrifuged at 600g for 3 min and resuspended in 10 ml of 0.168 M NH4Cl to lyse erythrocytes. After 10 min, cells were washed with 40 ml PBS and resuspended in 1 ml staining buffer (2% FCS, 1 mM EDTA, 25 mM HEPES-KOH pH 7.9 in PBS).
Cells were stained with the appropriate antibodies prior to purification by flow cytometry. For bone marrow pre-B cells, cell suspensions were stained with 6 μl each (6:1,000 dilution) of FITC anti-CD19 (BD Pharmingen, 553785) and PE anti-CD43 (BD Pharmingen, 553271). Bone marrow or spleen IgM+ cells were stained with 10 μl (1:100 dilution) FITC anti-IgM (BD Pharmingen, 553408) whereas spleen IgG+ cells were stained with 15 μl PE (3:200 dilution) anti-IgG (eBioscience, 12-4010-82). Following incubation at room temperature for 10 min, cells were washed with PBS and resuspended in 0.5 ml staining buffer prior to purification using a FACSMelody (BD) cell sorter running BDFACSChorus 3.0 software. CD19+/CD43− cells were isolated as the pre-B population whereas cells stained with anti-IgM+ or anti-IgG+ were isolated as their respective populations.
Patient samples
Patient samples, taken as part of routine diagnostics, were supplied by VIVO Biobank, HMDS or a hospital in the Czech Republic. Collection and use of patient samples were approved by the appropriate institutional review board (IRB). Each organization obtained informed patient consent for anonymized samples to be used by third parties for research. The use of surplus diagnostic material for research by HMDS and collaborators was approved by the Health Research Authority (HRA): 04_Q1205_125. Local ethics approval was obtained from the Biological Sciences Research Ethics Committee, University of Leeds: BIOSCI 18-031 & 2308 and CCR 2285, Royal Marsden Hospital NHS Foundation Trust.
Cell culture
hTERT-RPE-1 cells were from ATCC where they were authenticated by morphology, STR profiling and karyotyping and verified mycoplasma-free. NIH3T3 cells were from the laboratory of C. Bonifer, whereas HeLa cells were from the laboratory of T. Enver. Both cell types were authenticated using species-specific PCR primers. They were verified free from mycoplasma using MycoAlert Mycoplasma Detection Kit (LT07-318). HeLa cell contamination has caused misidentification of other cell lines. However, HeLa cells were used here only to prepare human DNA and we verified that the DNA was human using human-specific PCR primers. hTERT-RPE-1, NIH3T3 and HeLa cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum, 4 mM l-glutamine, 50 U ml−1 penicillin and 50 μg ml−1 streptomycin in a humidified incubator at 37 °C with 5% CO2.
Preparation of genomic DNA
Genomic DNA from mouse B cells, NIH3T3 cells, HeLa cells and patient samples was prepared by gently resuspending 1 × 106 to 5 × 106 cells in a 200 µl digestion buffer (200 mM NaCl, 10 mM Tris-HCl pH 7.5, 2 mM EDTA, 0.2% SDS). Proteinase K was added to a final concentration of 0.4 mg ml−1, followed by incubation at 56 °C overnight, with rotation. The next day, an equal volume of isopropanol was added and the sample mixed thoroughly but gently by inversion to precipitate the DNA. The DNA pellet was recovered by centrifugation at 20,000g for 5 min at room temperature and washed twice with 70% ethanol. DNA was then resuspended in 100 µl TE and incubated at 56 °C for at least 3 h to ensure complete resuspension; the concentration was measured using a DeNovix DS-11 spectrophotometer.
High-molecular-mass genomic DNA was prepared from fresh BCP-ALL bone marrow aspirates (that were surplus to diagnostic needs and that had been maintained at 4 °C), using a Promega Wizard HMW DNA extraction kit according to the manufacturer’s instructions for whole blood. Samples were resuspended in TE at room temperature overnight prior to measuring the concentration as above.
Isolation of total RNA and reverse transcription
Two million mouse B cells, NIH3T3 cells, HeLa cells or BCP-ALL cells were resuspended in 500 µl of TRIzol (Invitrogen, 3289) and total RNA was isolated according to the manufacturer’s instructions. DNA contaminants were removed by treatment with 2 U DNase I (Thermo Scientific, EN0521) for 45 min at 37 °C in 100 µl of 1× DNase I buffer (10 mM Tris-HCl pH 7.5, 2.5 mM MgCl2, 0.1 mM CaCl2). Following phenol-chloroform extraction and ethanol precipitation, total RNA was resuspended in 20 µl of ddH2O and the concentration was determined using a DeNovix DS-11 spectrophotometer.
One microgram of total RNA was reverse transcribed with M-MuLV reverse transcriptase (Invitrogen, 28025-013). In brief, 1 µg of RNA was added to 2.5 µM oligo dT primer, 500 µM dNTPs and ddH2O to give a total volume of 12 µl. This was incubated at 65 °C for 5 min and immediately placed on ice before addition of 4 µl first strand buffer (Invitrogen), 10 mM DTT and 1 µl RNase inhibitor (PCRBIO, PB30.23-02). The reaction was incubated at 37 °C for 2 min, followed by addition of 1 µl M-MuLV and incubation at 37 °C for 50 min prior to heat inactivation at 70 °C for 15 min.
Exonuclease V treatment of genomic DNA
Linear DNA was removed from genomic DNA using Exonuclease V (RecBCD, NEB M0345S). Reactions comprised 1 µg genomic DNA, 1× NEBuffer 4, 1 mM ATP and 10 U RecBCD in 100 µl total volume. Negative control reactions were identical except RecBCD was omitted. Following incubation at 37 °C for 1 h (mouse DNA) or 3 h (BCP-ALL DNA), EDTA was added to a final concentration of 11 mM and reactions were heat inactivated at 70 °C for 30 min. DNA was then ethanol precipitated and resuspended in 50 µl ddH2O; 5 µl (100 ng) was used directly for PCR.
Quantitative PCR
qPCR was performed using a Rotor-Gene 6000 cycler (Corbett) and analysed using the Corbett Rotor-Gene 6000 Series Software (v.1.7, build 87). All reactions were carried out in a final volume of 10 µl, containing 1× qPCRBIO SyGreen Mix (PCRBIO, PB20.14), 400 nM each primer and 100 ng genomic DNA, 1–5 ng cDNA or 1 µl first round PCR product (for nested PCR). All reactions were performed in duplicate. In each case, a standard curve of the amplicon was analysed concurrently to evaluate the amplification efficiency and to calculate the relative amount of amplicon in unknown samples. R2 values were 1 ± 0.1. A typical cycle consisted of: 95 °C for 3 min, followed by 40 cycles of 95 °C for 5 s, Tm for 10 s and extension at 72 °C for 10 s, where Tm = melting temperature of the primers (Supplementary Table 5). A melt curve, to determine amplicon purity, was produced by analysis of fluorescence as the temperature was increased from 72 °C to 95 °C. Amplicons were 100–200 bp.
Standard curves for absolute quantification were generated by 35 cycles of conventional PCR and purification of the desired product via a 1.2% agarose gel and a QIAquick gel extraction kit, (QIAGEN, 28704). Following measurement of the concentration via absorbance at 260 nm (using a DeNovix DS-11 spectrophotometer), DNA was diluted to 1–10 ng/μl before more accurate concentration determination using a QuantiFluor dsDNA kit (Promega, E2670) and a FLUOstar OPTIMA plate reader (BMG Labtech). An appropriate range of each standard was used in qPCR, ensuring that all unknown samples were within the standard curve.
Primers and melting temperatures are shown in Supplementary Table 5 for quantitative analysis of recombination, SJs and Rag1 expression in mouse bone marrow and spleen, and for quantitative (qPCR) analysis of recombination, SJ levels, RAG1 and RAG2 expression as well as PCNA, POLE3, POLE4 and RBX1 expression in BCP-ALL patient samples. BLAST was used to check primer specificity. HPRT was used as a reference gene for expression studies, using primers that span an intron. Genomic GAPDH sequences were used to normalize for cell numbers. These housekeeping genes were chosen for their widespread expression (HPRT) and low likelihood of mutation.
Detection of recombination in BCP-ALL patient samples
PCR was performed using Taq DNA polymerase (NEB, M0267) in reactions comprising 1× ThermoPol buffer, 200 µM dNTPs, 0.5 µM each primer, 1.25 U Taq DNA polymerase and 100 ng genomic DNA template in a final volume of 50 µl. Primers for recombination at the immunoglobulin kappa and lambda loci were as described by the BIOMED-2 consortium31. Cycling conditions involved initial denaturation at 95 °C for 5 min, followed by 95 °C for 30 s, 60 °C for 30 s and 68 °C for 30 s for 35 cycles, followed by a final extension of 5 min at 68 °C. PCR products were separated by gel electrophoresis; products of the expected sizes31 were excised and cloned prior to Sanger sequencing.
Nested PCR of recombination junctions and SJs in mouse and patient samples
To achieve sufficient specificity and sensitivity, nested PCR was performed using Taq DNA polymerase (NEB, M0267). First round reactions consisted of 1× ThermoPol buffer, 200 µM dNTPs, 0.5 µM each primer, 1.25 U Taq DNA polymerase and 20–100 ng genomic DNA template in a final volume of 50 µl. To detect mouse recombination and SJs, thermocycling conditions involved denaturation at 94 °C for 3 min, followed by 18 cycles of 94 °C for 20 s, Tm for 20 s and 72 °C for 20 s. A 7 min of final extension was performed. One microlitre of 1:10 diluted first round PCR product was used as the template for a second round of qPCR using the primers shown in Supplementary Table 5.
As the BIOMED-2 primers to detect human SJs do not robustly identify specific J gene segments, nested PCRs were carried out with a mixture of first round primers that bind upstream of all five KJ RSSs or LJ1, LJ2 and LJ3 RSSs, which account for ~99% of IGL recombination events31, followed by second round PCRs specific for each individual J RSS. First round reactions were set up as described above with 50–100 ng template DNA, followed by denaturation at 95 °C for 5 min, and 25 cycles of 95 °C for 30 s, Tm for 30 s and 68 °C for 30 s, and a final extension of 5 min at 68 °C. Second round reactions were identical except 1 µl first round PCR product was used as template and the number of cycles was optimized for each amplicon, which was typically 36 cycles. Primers and melting temperatures are shown in Supplementary Table 5.
Sequencing of PCR products
PCR products were separated on a 1.2% agarose gel; DNA was purified from the gel using a QIAquick gel extraction kit (QIAGEN, 28704) and cloned into a T-tailed pBluescript II SK (+) vector (Stratagene, 212205) that had been digested with EcoRV-HF (NEB, R3195). Positive clones were sent for Sanger sequencing (Eurofins Genomics, LightRun Tube) using a M13 forward sequencing primer.
Sequencing traces (in .ab1 format) were aligned using SnapGene (v4; GSL Biotech). For recombination, sequences were aligned against the human immunoglobulin kappa (NCBI Gene ID: 50802) or lambda loci (NCBI Gene ID: 3535), as appropriate. For ESCs, the possible head-to-head SJ sequence was assembled from the appropriate genomic sequence and sequences were aligned to this. All alignments were verified by BLASTN (NCBI; accessed at https://blast.ncbi.nlm.nih.gov), where the search set was limited to the Homo sapiens (taxid: 9606) RefSeq Genome Database (refseq_genomes).
ddPCR
ddPCR reactions were conducted in a total volume of 20 µl with 1× ddPCR Supermix for probes (Bio-Rad 1863026), 900 nM of each primer, 250 nM probe and 63 ng template DNA. Droplets were generated in an 8-well droplet generation plate using a Bio-Rad QX100 droplet generator. Nanodroplets were carefully transferred to a 96-well plate, which was sealed with foil prior to thermocycling. The latter involved an initial denaturation at 95 °C for 10 min, followed by 40 cycles of 94 °C for 30 s and 60 °C for 1 min, followed by 98 °C for 10 min and 4 °C for 30 min to allow droplets to equilibrate. The presence of amplified products was determined using the Bio-Rad QX100 Droplet Reader and QuantaSoft v1.7.4 software. For positive droplet identification, a manual threshold (2000) was applied to 1D amplitude data to minimize the occurrence of false positives. Primers are shown in Supplementary Table 5, for absolute quantitative analysis of GAPDH, recombination junctions and SJs in BCP-ALL samples.
Targeted sequencing of recombination and SJs (LAM-ESC and LAM-recombination)
Targeted sequencing of light chain recombination and SJs was performed using modified versions of the LAM-HTGTS technique32 with bespoke analysis pipelines. Recombination junctions were detected using LAM-recombination, where bait primers (Supplementary Table 5) were designed against regions adjacent to J gene segments, allowing recombination to V gene segments to be determined. SJs were detected via LAM-ESC where bait primers (Supplementary Table 5) were designed against regions adjacent to J segment RSSs, allowing any sequence (for example, V RSSs) joined to J RSSs to be determined.
Libraries were generated as described32 with minor modifications. Specifically, 500 ng genomic DNA was used as template in 90 cycles of the initial Bio-PCR. For the final PCR step (Tagged PCR), primers were used to add sequencing adaptors (Amplicon-EZ-I7-blue and Amplicon-EZ-I5-nested; Supplementary Table 5). Following library generation, samples were sent for 2× 250 bp paired-end sequencing using the Amplicon-EZ service (Azenta).
To analyse the data, FASTQ files were initially demultiplexed using each J gene segment or J RSS nested primer, for recombination and SJ libraries, respectively. The paired-end reads were then combined into a single read, using the overlap at the 3′ end of read 1 and 5′ end of read 2. If reads could not be combined, read 1 only was analysed. A custom Python script was used to automate BLAST searches against a custom BLAST database, consisting of all V-J recombination events or all head-to-head RSS combinations from the immunoglobulin kappa and lambda loci, for recombination and SJ libraries, respectively https://github.com/Boyes-Lab/LAM-ESC-Recombination33.
Clonotype determination
Recombination junctions were amplified using LV3-1_REC_F, LV5-45_REC_F or LV2-11_REC_F with LJ2/3_REC_R (Supplementary Table 5). Following gel purification of the amplified products using the QIAquick gel extraction kit (QIAGEN, 28704), samples were sent for 2 ×250 bp paired-end sequencing using the Amplicon-EZ service (Azenta) that included addition of Amplicon-EZ-I7 and Amplicon-EZ-I5 sequencing adaptors. Paired-end reads in the .fastq format were combined by overlapping the 3′ end of read 1 with the 5′ end of read 2 (EGAD50000001518). The reads were compared to the reference motifs near the breakpoint junction of interest using the script Clonotype_analysis.py (https://github.com/Boyes-Lab/NGS-Analysis)48, which identified each unique clonotype and the frequency at which it occurred. The reference motifs consisted of 5 bp from each of the respective V and J motifs, derived from sequences that lie 20 bp from the V-J junction that would be formed in the absence of processing. Specifically, the reference motifs were: 5 bp of V gene reference sequence; omit 20 bp of V gene sequence to the boundary; omit 20 bp of J gene sequence; use 5 bp of J gene sequence as reference. If the amplified sequence contained both of the 5bp motifs, then the code identifies each unique sequence that intervenes. The identified sequences were then inputted into IgBlast55 which determined if the insert at the V-J junction was derived from elsewhere in the genome. The number of unique clonotypes was determined from these inserts: two sequences with the same V-J insert were classed as being the same clonotype.
The number of recombination copies attributable to each distinguishable minor clonotype was determined by calculating the absolute copy number of the recombination event by ddPCR and multiplying that by the percentage of each minor clonotype. Assuming each recombination is present on a single allele, the number of cells harbouring the recombination event was then estimated (that is, one recombination copy equates to one cell harbouring the recombination). The number of cell divisions (n) required to generate the recombination levels measured for each minor clonotype was subsequently calculated using the formula for cell population doublings: N2 = N1(2n) where N1 = number of cells at beginning (that is, 1 cell) and N2 = number of cells at end (that is, the estimated number of cells). From this, the minor clonotypes were calculated to be present at copies equivalent to 0.81–3.69 relative cell divisions (patients R-8 and R-13).
To investigate if the SJ levels observed via ddPCR (Extended Data Fig. 7c) could have resulted from replication of SJs from the minor clonotypes, the number of SJs predicted to remain if ESCs are diluted at each cell division were calculated using the formula for exponential decay: xt = x0/2t where xt = predicted SJ level, x0 = initial SJ level (that is, 1) and t = number of cell divisions (calculated above). Theoretically, the SJ measured by ddPCR could correspond to one or more of the minor clonotypes; we therefore took a conservative approach and summed the predicted SJ values for all the distinguishable clonotypes. We then divided the observed SJ value (Extended Data Fig. 7c) by the predicted SJ levels for the minor clonotypes. This observed/predicted value was compared to the observed/predicted values for SJs generated a similar number of relative cell divisions ago, using the values shown in Fig. 3c, where the SJs were estimated to result from 0.33–6.32 relative cell divisions. The SJ copies found in patient R-8 are substantially higher than those generated by replication of SJs from similarly recent recombination events (Extended Data Fig. 7d), implying that at least some of the SJs have persisted from the primary recombination event. Similar analyses cannot discount the possibility that replication of recently generated SJs generated the SJs observed in patients R-13 and R-9.
Calculation of observed/predicted ESC levels
The observed SJ/predicted SJ ratio provides a more accurate measure of ESC replication as it takes the extent of cell division into account. It ensures that ESCs from cells that have undergone marked differences in cell division do not artefactually show the same level of replication. Predicted SJ levels were calculated using the formula for exponential decay given above. For Fig. 3c, only recombination junctions that had undergone ≤6 cell relative divisions per ddPCR sample were examined.
Phi29 amplification
Freeze-thawing of cells causes DSBs56,57 and depletion of circular DNAs in patient samples. Phi29 was therefore used to amplify remaining circular DNAs in BCP-ALL samples via rolling circle replication, using the Illustra TempliPhi 500 Amplification kit (Cytiva 25640010) according to the manufacturer’s protocol. Specifically, 10 ng of DNA was diluted with 50 μl of sample buffer, incubated at 95 °C for 3 min and cooled to 4 °C. The reaction was then incubated in 1× Phi29 reaction buffer (50 mM Tris-HCl pH 7.5, 10 mM MgCl2, 10 mM (NH4)2SO4, 4 mM DTT) with 2 μl of enzyme mix at 30 °C for 18 h (human DNA) or 6 h (mouse DNA), followed by 65 °C for 10 min to inactivate the enzyme. DNA was precipitated with isopropanol, washed with 70% ethanol and resuspended in 25 μl ddH2O. Control experiments omitted the enzyme. Sample concentrations were determined via absorbance at 260 nm (using a DeNovix DS-11 spectrophotometer) and diluted to 7 ng/µl. SJs were quantified by ddPCR and the SJ/GAPDH ratio of treated versus untreated sample determined.
Fluorescence in situ hybridization
DNA-FISH was performed as described58. Fosmid clones targeting IGK (ABC10-44246300H4), IGK-JK-KDE (ABC8-2123240B1) and IGL (ABC10-44455600K21) ESCs as well as IGK and IGL control regions (ABC10-43608900D2 and ABC10-44444000A2, respectively) were gifts from E. Eichler. Each fosmid probe was directly labelled by nick translation as described59, except the amount of DNA labelled was reduced to 1 μg and both aminoallyl-dUTP and aminoallyl-dCTP were incorporated, followed by coupling to fluorescent dyes (Alexa Fluor 488/555/647, Invitrogen). Fosmid probes were purified using a QIAquick PCR purification kit and elution in 10 mM Tris-HCl pH 8.5. Patient bone marrow samples were cultured in StemSpan SFEMII medium (Stem Cell Technologies) supplemented with 20% fetal calf serum, 1% l-glutamine, 100 μg ml−1 Primocin (InvivoGen), 20 ng ml−1 IL-3 and 20 ng ml−1 IL-7 (Cell Guidance Systems) in a 37 °C humidified incubator, 5% CO2, prior to Colcemid treatment (0.2 µg ml−1, KaryoMAX, ThermoFisher) for 2 h. hTERT-RPE-1 cells were maintained in supplemented DMEM and treated with Colcemid as described above. Cells were then centrifuged and resuspended in prewarmed 75 mM KCl, followed by incubation at 37 °C for 20 min. After further centrifugation, cells were resuspended in Carnoy’s fixative (methanol: glacial acetic acid 3:1) and dropped onto humidified microscope slides. Slides were incubated in 2× SSC/RNase A 100 µg ml−1 at 37 °C for 1 h, followed by successive dehydration in 70%, 90%, and 100% ethanol. Slides were heated to 70 °C for 5 min on a hot plate, followed by DNA denaturation in preheated denaturant (2× SSC/70% formamide) at 70 °C for 30 min. Subsequently, slides were placed in ice-cold 70% ethanol, then 90% and 100% ethanol at room temperature before air-drying. For each slide, 100 ng of fosmid probe was combined with 6 µg human Cot-1 DNA (Invitrogen) and 5 µg single stranded DNA from salmon testes (Invitrogen), followed by ethanol precipitation. The DNA pellet was washed with 70% ethanol and resuspended in hybridization buffer (2× SSC, 50% deionized formamide, 10% dextran sulfate, 1% Tween-20). FISH probes were denatured at 92 °C for 5 min, pre-annealed at 37 °C for 15 min and then were immediately hybridized with DNA on slides overnight at 37 °C in a light-tight humidified chamber. Slides were washed in 2× SSC at 45 °C, 0.1× SSC at 60 °C, 1× PBS with 10 µg ml−1 DAPI at room temperature and finally mounted with SlowFade Gold antifade reagent (Invitrogen). Slides were imaged using an Olympus IX83 widefield fluorescence microscope with a 60× (60×/1.4 Oil, Plan Apo (oil)) objective and a Photometrics Prime BSI CMOS camera with a motorized xyz stage. Filter sets are DAPI (excitation 365/10 nm) emission 440/40 nm, GFP (excitation 482/24 nm) emission 530/40 nm, RFP (excitation 545/10 nm) emission 600/50 nm, Cy5/A647 (excitation 628/40 nm) emission 692/40 nm. Images were acquired using Olympus CellSens Dimension 3.2 (Build 23706) software and analysed using FIJI 2.16.0 software.
BrdU immunofluorescence
Bone marrow cells were resuspended in StemSpan SFEMII medium, supplemented as described above, and labelled with 10 µM BrdU (Merck B5002) for 28 or 48 h in a 37 °C humidified incubator, 5% CO2. Primary BCP-ALL cells60 have a doubling time of 26 to 240 h. Therefore, for cells incubated with BrdU for 28 h, incorporation should be limited to a single S phase for any cells in metaphase. Chromosome spreads were prepared as described for DNA-FISH. DNA was denatured by incubating the slides in 1 M HCl for 40 min at room temperature, followed by neutralization in 0.1 M Borate buffer pH 8.5 for 15 min at room temperature. Slides were then washed in 0.1% Triton X-100/PBS and blocked in 0.1% Triton X-100/PBS/5% goat serum (Merck G9023) for 1 h at room temperature. Following immunostaining with a 1:500 dilution of anti-BrdU antibody (BD Pharmingen, 555627) for 1 h at room temperature, cells were incubated with goat anti-mouse secondary antibody (Jackson ImmunoResearch, 115-001-003) at a 1:1,000 dilution for 1 h at room temperature, followed by incubation with Alexa Fluor Plus 488 labelled donkey anti-goat antibody (ThermoFisher, A32814), also at 1:1,000 dilution for 1 h at room temperature. Following counterstaining with DAPI (10 µg ml−1, Invitrogen), images were captured using an Olympus IX83 widefield fluorescent microscope and CellSens software and analysed using FIJI 2.16.0 software as above.
PCR amplification of the SYK gene
PCR was performed using Taq DNA polymerase (NEB, M0267) using the conditions described above for BIOMED-2 primers except the final volume was 25 µl. Primers are given in Supplementary Table 5. Cycling conditions involved initial denaturation at 95 °C for 3 min, followed by 95 °C for 20 s, 64 °C for 30 s and 68 °C for 30 s for 39 cycles, followed by a final extension of 5 min at 68 °C. PCR products were separated by gel electrophoresis.
Analysis of ETV6–RUNX1 BCP-ALL WGS data for ESCs
WGS datasets from patients with BCP-ALL (downloaded from the European Genome-phenome Archive (EGA), dataset ID: EGAD00001000116) were analysed for SJs using an in-house Python script (https://github.com/Boyes-Lab/NGS-Analysis)48. In brief, paired-end sequencing data was aligned to the hg19 build of the human genome using Bowtie2 in local alignment mode. Following alignment to the genome, the data were filtered for discordant reads at the immunoglobulin and TCR loci using Samtools. Reads were further filtered (using the above in-house Python script) to extract divergent reads, indicative of SJs. Similar tools that capitalize on discordant paired-end reads have been developed to map ecDNAs. However, AmpliconArchitect40 also requires increased circular DNA copy number whereas CircleSeq involves removal of linear DNA prior to sequencing and analysis44,61. AmpliconArchitect40 did not detect circular DNAs in WGS from 20 patient samples where ESCs were detected by LAM-ESC (Fig. 2; EGA accession code: EGAS00001006863). This is likely because unlike ecDNAs, individual ESC sequences do not undergo copy number amplification that is required for detection by AmpliconArchitect40.
Identification of differentially expressed genes and GSEA analysis
RNA-seq data of patients at diagnosis were downloaded from the TARGET database (dbGaP Sub-study ID: phs000464; Fig. 3d) or EGA (Accession Code EGAS00001006863; Fig. 3f). Differentially expressed genes were identified using DESeq2 with |logFC| > 0.585 and FDR < 0.05. Adjustment for multiple comparisons was performed via the DESeq2 programme. Gene set enrichment analysis (GSEA) was carried out according to the user guide provided by the BROAD Institute (https://docs.gsea-msigdb.org/#GSEA/GSEA_User_Guide/).
Whole-exome sequencing
DNA was prepared from germline and BCP-ALL patient samples taken at diagnosis and relapse, using the Promega Wizard HMW DNA extraction kit. The concentration was measured via Qubit and whole-exome paired-end read sequencing was performed at 100× depth by Azenta.
Analysis of SVs
SV data of patients at diagnosis and relapse were downloaded from Complete Genomics (CGI, from within the TARGET database: dbGaP Sub-study ID: phs000464; Fig. 5a,b). For Extended Data Fig. 9d, WGS of patients with BCP-ALL in the VIVO Biobank cohort was downloaded from EGA under the Accession Code EGAS00001006863. The analysis used similar numbers of WGS and WES where WES from germline and BCP-ALL samples are available from EGA (dataset ID: EGAD50000001519). Breakpoints of SVs were analysed using a bespoke Python program, SVs_near_RSSs.py, which creates an analysis window spanning 50 bp either side of each breakpoint (https://github.com/Boyes-Lab/Structural-Variants)62. The presence of an RSS within the window is then analysed using the DNAGrep algorithm, via RSSsite63.
The relative impact of cut-and-run, ESC reintegration and RAG-mediated insertion was determined as follows: SVs were defined as either deletions, insertions, translocations or complex insertions based on the source of the DNA strand on either side of an SV breakpoint. The DNA damage occurring near RSS sites was analysed, as RAG-mediated DSBs are required for both reintegration and cut-and-run-mediated DNA damage. ESC reintegration events are defined by the reinsertion of ESC DNA into the genome. Thus, insertions were considered to be reintegration-derived where the inserted DNA came from immunoglobulin or TCR loci. The proportion of these events near RSS sites was then compared with other types of DNA damage. Open-source scripts to analyse potential cut-and-run events compared to reintegration are provided at https://github.com/Boyes-Lab/Structural-Variants62.
Analysis of ecDNAs using AmpliconArchitect40
In silico experiments to detect ecDNAs were carried out using AmpliconArchitect40 according to the AmpliconSuite pipeline (https://github.com/AmpliconSuite/AmpliconSuite-pipeline/blob/master/documentation/GUIDE.md) using WGS BAM files from VIVO Biobank patient samples available from EGA (Accession Code EGAS00001006863).
Statistics
All statistical analyses were performed using GraphPad Prism v9. Statistical test results are provided as P values in the figures. Detailed descriptions of error bars and the number replicates and/or cells analysed are reported in the figure legends. Biological replicates are shown unless otherwise indicated. Analyses of fold changes between biological replicates were performed using a two-tailed Student’s t test or Mann–Whitney U test (depending on data distribution) where *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. The 95% confidence intervals are given in the figure legends where possible. The 95% confidence intervals for the difference of mean gene expression (H-relapse versus non-relapse) in Fig 3e: PCNA: −2.007 to −0.1691; POLE3: −2.206 to −0.1488; POLE4: −4.910 to −0.8914; RBX1: −2.206 to −0.6157. The Kolmogorov–Smirnov test was used to determine whether the data followed a Gaussian distribution. GSEA uses the statistical test described53,54 with corrections for multiple comparisons. DeSeq2 analysis used the Wald statistical test with the Benjamini–Hochberg correction for multiple testing. Statistical analyses with two categorical variables were performed using a two-way ANOVA. Statistical analyses of the proportion of ESCs above the threshold in patients who relapse versus those who do not were determined using a two-tailed Fisher’s exact test. The significance of the difference between matched ESCs in relapse and non-relapse groups was determined using a two-tailed Wilcoxon signed-rank test. Pearson correlation coefficients (r values) were computed for scatter plots and tested (null r = 0) with a standard two-tailed test. The significance of SVs involving single cRSSs occurring more frequently at relapse-associated genes was calculated using the hypergeometric distribution (Fig. 5b and Extended Data Fig. 9d) to analyse the number of SVs involving single cRSSs within relapse-associated genes compared to SVs involving single cRSSs in the whole dataset. The significance of the co-localization of breaks detected by LAM-HTGTS and genes that are frequently mutated in relapsed ALL (Fig. 5c) was calculated by using the hypergeometric distribution (implemented in the R software, https://www.r-project.org) to test whether the number of genes which were more commonly mutated with SJ partner versus 12-RSS or 23-RSS partners (controls) occurred more frequently in the relapse-associated genes versus the whole gene list. A power calculation, taking an alpha value of 0.05 and a desired power of 80% was used to initially estimate the sample size in which to analyse ESC levels.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.