Animals
All animal experiments conformed to the Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Committee of Laboratory Animal Experimentation of the RIKEN Center for Biosystems Dynamics Research. B6D2F1 (C57BL/6âÃâDBA/2) and C57BL/6 mice, aged 8â10 weeks, were used to produce oocytes and sperm. For allele-specific analysis, C57BL/6 female mice and MSM/Ms male mice were used to produce oocytes and sperm, respectively. To eliminate the effect of individual differences between mice, multiple mice were used in each experiment as follows. Figure 1b, 2 (4-cell), 4 (8-cell), 2 (16-cell) and 4 (ICM and TE) mice; Fig. 1d, 10 mice each (1-cell, 2-cell and 4-cell); Fig. 2d, 4 (1-cell), 3 (2-cell) and 3 (4-cell) mice; Fig. 2f, 3 (1-cell), 3 (2-cell) and 4 (4-cell and 8-cell) mice; Fig. 3b, 4 in vitro and 2 in vivo (2-to-4-cell), 8 in vitro and 7 in vivo (4-to-8-cell), and 4 in vitro and 2 in vivo (8-to-16-cell) mice; Fig. 3c, 15 mice each (2-to-4-cell, 4-to-8-cell and 8-to-16-cell); Fig. 3g, 3 mice each (2-to-4-cell, 4-to-8-cell, 8-to-16-cell and 16-to-32-cell); Fig. 3h, 4 (2-cell) and 3 (4-cell and 8-cell) mice; Fig. 3i, 5 (2-cell, 4-cell, and 8-cell) mice; Fig. 4d, 4 mice; Fig. 4hâi, 6 mice; Fig. 4j, 16 mice; Extended Data Fig. 7a, 7 (2-to-4-cell DMSO), 6 (2-to-4-cell aphidicolin 15ângâmlâ1, 30ângâmlâ1, and 60ângâmlâ1), 8 (2-to-4-cell aphidicolin 75ângâmlâ1), 16 (4-to-8-cell DMSO), 6 (4-to-8-cell aphidicolin 15ângâmlâ1, 30ângâmlâ1 and 60ângâmlâ1), 6 (4-to-8-cell aphidicolin 75ângâmlâ1), 12 (8-to-16-cell DMSO), 8 (8-to-16-cell aphidicolin 15ângâmlâ1, 30ângâmlâ1 and 60ângâmlâ1) and 6 (8-to-16-cell aphidicolin 75ângâmlâ1) mice.
Oocyte and embryo collection
The temperature, humidity and light cycle of mouse cages were maintained at 20â24â°C, 45â65% and 12âhâ12âh darkâlight, respectively. Mature oocytes were collected from the oviducts of eight- to ten-week-old female mice that had been induced to superovulate with 5âIU of equine chorionic gonadotropin (eCG, ASKA Pharmaceutical) followed by 5âIU of human chorionic gonadotropin (hCG; ASKA Pharmaceutical) 48âh later. Cumulus-oocyte complexes were collected from the oviducts approximately 16âh after hCG injection. Cumulus-oocyte complexes were placed in M2 medium and treated with 0.1% (w/v) bovine testicular hyaluronidase. After several minutes, the cumulus-free oocytes were washed twice and then moved to Chatot, Ziomek and Bavister medium (CZB). Mature metaphase II (MII) oocytes were subjected to in vitro fertilization (IVF), intracytoplasmic sperm injection (ICSI) or SCNT. Developmentally arrested or delayed embryos at each embryonic day were excluded from further analysis. In vivo developed zygotes and embryos were collected from the oviducts of pregnant mice that had been induced to superovulate and mated with male mice. For in vitro development, in vivo developed zygotes were cultured in CZB. As we pooled embryos from multiple females before single-embryo sampling, our dataset is not suitable for female-to-female variability assessment. When indicated, embryos were cultured in the presence of nucleosides (Sigma-Aldrich, EmbryoMax Nucleosides, 100Ã; at a final concentration of 2Ã) or aphidicolin (FUJIFILM Wako). In Extended Data Fig. 7a,b, embryos were monitored under the microscope every 30âmin, and embryos that had just completed cell division were judged to be in G1 and transferred to the medium containing aphidicolin. The subsequent M phase was monitored by live imaging as in Extended Data Fig. 7a. Embryos that had just completed the 4-to-8-cell division were collected for scRepli-seq as in Extended Data Fig. 7b.
Fertilization
IVF was performed according to the manufacturerâs instructions using CARD MEDIUM (Cosmo Bio). To limit fertilization, we set the insemination time to 1âh. Intracytoplasmic sperm injection with sperm heads was performed as described previously61. In brief, the sperm head was separated from the tail by applying several piezo pulses to the neck region, and the head was then injected into an oocyte. After 20âmin of recovery at room temperature, injected oocytes were cultured in CZB.
Single-cell (blastomere) collection
Collection of single blastomeres was performed using micromanipulation. Embryos were transferred into M2 medium supplemented with 5âμgâmlâ1 cytochalasin B (Sigma-Aldrich) for 10âmin. The zona pellucida was then cut using the LYKOS laser system (Hamilton Thorne) in a micromanipulation chamber, which was placed onto a warmed stage (37â°C) in an inverted microscope (Olympus). After cutting the zona pellucida, a fire-polished injection pipette (inner diameter, 30âµm) was inserted through the hole of the zona pellucida, and single blastomeres were isolated. For late-8-cell- and 16-cell-stage embryos, embryos were treated with TrypLE Express (Gibco) for 5âmin before single-cell isolation. TE and ICM cells were isolated as previously described62. Microsurgical isolation of single cells was performed by micromanipulation. After single-cell isolation, the cell was washed twice with PBS. The cell in 0.5âµl PBS was transferred to a tube with 6âµl sampling buffer.
SCNT
SCNT was performed as previously described61. In brief, groups of MII oocytes were transferred into droplets of M2 medium containing 500âµgâmlâ1 cytochalasin B on the microscope stage to collect the MII spindle. Oocytes undergoing microsurgery were held with a holding pipette. A hole was made in the zona pellucida through the application of several piezo-pulses using an enucleation pipette. The MII spindle was aspirated into the pipette with a minimal volume of ooplasm, and the resulting enucleated oocytes were transferred into CZB. For nuclear injection, donor cumulus cells were gently aspirated in and out of the injection pipette to broken plasma membranes. Each nucleus was injected into an enucleated oocyte, and these reconstructed oocytes were kept in the incubator until activation. Reconstructed oocytes were parthenogenetically stimulated by incubation in CZB supplemented with 10âmM SrCl2, 2âmM ethylene glycol tetraacetic acid (EGTA), and 5âµM latrunculin A for 10âh, and then cultured in CZB.
Single-cell DNA replication profiling (scRepli-seq) using mouse embryos
scRepli-seq experiments using mouse embryos were performed as previously reported with slight modifications. In brief, unfixed single blastomeres (derived from BDF1 or B6MSM strain embryos) were collected into 8-well PCR tubes with 6âμl cell lysis buffer (288âμl of H2O, 2âμl of 10âmgâmlâ1 proteinase K (Sigma-Aldrich, P4850), 32âμl of 10à single-cell lysis and fragmentation buffer (Sigma-Aldrich, L1043)), incubated at 55â°C for 1âh and then at 99â°C for 4âmin for gDNA isolation and fragmentation. For scRepli-seq analysis, we analysed all blastomeres of an embryo unless there was accidental damage to the cell/sample. For scRepli-seq experiments after EdU staining (Extended Data Fig. 3b,c), after taking the photographs of EdU-stained cells (according to the protocol in Extended Data Fig. 3a), the cells were manually collected by a mouth pipette under the microscope into 12âμl of cell lysis buffer and incubated at 55â°C for 16âh (not 1âh). The remaining whole-genome amplification process (SeqPlex enhanced DNA amplification kit, Sigma-Aldrich, SEQXE) and next-generation sequencing (NGS) library construction (NGS LTP library preparation kit, KAPA, KK8232) were basically performed according to the manufacturerâs instructions. The samples were processed for NGS on the Illumina Hiseq 1500 or Hiseq X Ten system (80-bp-length single-read or 150-bp-length paired-end read sequencing).
Immunostaining
Embryos were fixed with 2% paraformaldehyde in PBS-polyvinyl alcohol (PVA) (pHâ7.4) for 30âmin. After blocking and permeabilization in PBS-PVA containing 1âmgâmlâ1 BSA (PBS-PVA-BSA) and 0.1% Triton X-100, the embryos were incubated with appropriate primary antibodies overnight at 4â°C, washed several times in PBS-PVA-BSA and incubated with secondary antibodies for 90âmin at room temperature. DNA was counterstained with 40âµgâmlâ1 of Hoechst 33342. Finally, the embryos were washed and transferred to BSA-PVA for imaging with a Zeiss LSM780 confocal microscope. The following primary antibodies were used: mouse anti-γH2A.X (phosphorylated Ser139) (1:200, Abcam, ab22551); rabbit anti-histone H3 (1:200, Abcam, ab62706); mouse anti-histone H3 (1:200, Abcam, ab195277); mouse anti-histone H3K9me2 (1:200, Monoclonal Antibody Institute, Japan (MABI), MABI0317); rabbit anti-phosphorylated-CHK1 (Ser345) (133D3) (1:200, Cell Signaling Technology, 2348S) antibodies. The secondary antibodies were Alexa Fluor 488 goat anti-mouse IgG (H+L) (A11029); goat anti-rabbit IgG (H+L) (A11034); Alexa Fluor 555 goat anti-mouse IgG (H+L) (A21424) (1:400, Invitrogen).
Quantification of fluorescence signals
To quantify the levels of H3K9me2, p-Chk1 or γH2AX relative to the levels of histone H3, we obtained the mean signal intensity for H3K9me2, p-CHK1 or γH2AX within the nuclei (Ime_nuc). We then subtracted the mean cytoplasmic signal intensity (Ime_cyto), which was obtained from a region near the nuclei, from the Ime_nuc value (Ime_nucâââIme_cyto). Similarly, we determined the histone H3 level within the same nuclei (IH3_nucâââIH3_cyto). Finally, we calculated the ratio between the two values (Ime_nucâââIme_cyto)/(IH3_nucâââIH3_cyto).
EdU staining
For each embryo, the timing of its fertilization or cleavage was recorded by observation using stereomicroscopy every 30âmin. At each hour after fertilization or cleavage, embryos were collected, treated with 20âµM EdU for 30âmin, and then fixed in 3.7% paraformaldehyde in PBS-PVA (pHâ7.4) for 30âmin. EdU staining was performed using Click-iT Plus EdU Alexa Fluor 555 or 594 Imaging Kit (Invitrogen). After EdU staining, samples were incubated with mouse anti-histone H3 (1:200, Abcam, ab195277) primary antibody at 4â°C overnight, washed several times in PBS-PVA-BSA and incubated with Alexa Fluor 488 goat anti-mouse IgG (H+L) (A11029) secondary antibody for 120âmin at room temperature. The embryos were finally washed and transferred to BSAâPVA for imaging on the Zeiss LSM780 confocal microscope. The images were reconstructed into 3D with Imaris software. In Fig. 2bâd and Extended Data Fig. 3aâc, we categorized EdU staining patterns based on spatial distribution and intensity. Images of nuclei were subjected to auto-thresholding with the âdefaultâ algorism in Fiji software, and the thresholded patterns were manually categorized into âuniformâ, ânuclear periphery + NPBsâ, âNPBsâ, ânPBs + internal fociâ, ânuclear periphery + internal fociâ, and âinternal fociâ. The âuniformâ category was further divided into two groups: those with nuclear EdU intensity 1.5à higher relative to cytoplasmic intensity (strong) and the others (weak). If the nuclear EdU intensity was less than 1.1 times the cytoplasmic intensity, it was categorized as âno signalâ. For the experiment shown in Extended Data Fig. 3a, EdU-stained cells after Hoechst 33342 treatment (20âμM, for 30âmin at 37â°C) were analysed using fluorescence-activated cell sorting using the Sony SH800 cell sorter using the single-cell mode (SH800 v.2.1).
EdU staining of metaphase chromosome spreads
The MC12 cells (cultured in 10% FBS/DMEM medium) and embryos (1-cell, 2-cell and 4-cell; C57BL/6 strain) were labelled with 20âμM EdU for 60âmin in the presumptive early-S phase. After several hours of cultivation without EdU, the cells and embryos were treated with 0.01âμgâmlâ1 colcemid for 120âmin to synchronize to M phase. The cells and embryos, from which the zona pellucida was removed by acidic Tyrodeâs solution, were exposed to hypotonic 1% FBS/PBS supplemented with 0.075âM KCl for 10â20âmin. The cells and embryos were then fixed with 3:1 methanol:acetic acid solution at â20â°C for 30âmin (cells) or room temperature for a few minutes (embryos), and the chromosome spreads were prepared on glass slides. EdU staining of metaphase chromosomes was performed using the Click-iT EdU Cell Proliferation Kit (Invitrogen), and EdU fluorescence signal intensity was analysed with ImageJ.
DNA fibre spreading assay
More than 30 embryos (C57BL/6 strain) at each developmental stage were collected. 1-, 2- and 4-cell embryos were collected at early S phase. Likewise, most 8-cell embryos were also collected at early-S phase (although some cells may not be at early S, as cells lose cell cycle synchrony after the 8-cell stage). Embryos were labelled with 100âμM 5-iodo-2â²-deoxyuridine (IdU) for 30âmin just before collection. After three quick washes with KSOM medium, the embryos were labelled with 100âμM 5-chloro-2â²-deoxyuridine (CldU) for 30âmin. The labelling reaction was stopped by washing the cells with ice-cold 1% FBS/PBS. The embryos, from which the zona pellucida was removed by acidic Tyrodeâs solution, were transferred into 1% FBS/PBS under the microscope with a mouth pipette. The embryos were then placed onto an APS-coated glass slide (Matsunami) with less than 1âμl of 1% FBS/PBS. Then, 20âμl of spreading buffer (0.5% SDS, 200âmM Tris-HCl (pHâ7.5), 50âmM EDTA, 100âmM NaCl) (NaCl was added for longer fibre recovery63) was added onto the embryos on the glass slides, which were incubated at room temperature for 6âmin (ref. 9). The slides were then gently tilted 20° from horizontal to stretch the DNA fibres. The DNA fibre slides were dried at room temperature for at least 1âh and the slides were fixed (methanol:acetic acid, 3:1) for 2âmin. DNA fibres on the fixed slides were denatured with 1âM NaOH for 22âmin, neutralized by five washes with PBS, and blocked with 1% BSA/PBST (0.05% Tween-20). Immunostaining of labelled DNA was performed with mouse anti-BrdU antibody (1:5; recognizes IdU and CldU64; BD, 347580) and rat anti-BrdU antibody (1:25; recognizes CldU64; Abcam, 6326) at 37â°C for 45âmin followed by incubation with Alexa Fluor 488 donkey anti-mouse IgG (H+L) secondary antibody (1:200, Invitrogen, A21202) and Alexa Fluor 594 donkey anti-rat IgG (H+L) secondary antibody (1:200, Invitrogen, A21209) at 37â°C for 30âmin. After these antibody incubation steps for IdU/CldU detection, the slides were further incubated with mouse monoclonal antibody against ssDNA (1:100, Millipore, MAB3034, 16-19) for 30âmin at 37â°C and Alexa Fluor 647 goat anti-mouse IgG2a secondary antibody (1:50, Invitrogen, A21241) for 30âmin at 37â°C to avoid cross-reaction65. Finally, the samples were mounted with ProLong Diamond (Invitrogen, P36970), and photographs were taken with the DeltaVision Elite microscope using a Ã60 Plan/Apo NA1.42 oil-immersion objective at 2,048âÃâ2,048âpixels. The excitation and emission band-pass filter sets used were 542/27 and 594/45ânm, respectively (TRITC), or 632/22 and 679/34ânm, respectively (Cy5), to avoid signal overlap between Alexa Fluor 594 and 647. Using λDNA as a control, we estimated the extension rate to be around 4.4âkb per μm. As the length of the smallest individual dots (30âmin labelled DNA) that we could observe by imaging using the DeltaVision microscope was approximately 0.3âμm (~1.3âkb), the maximum resolution of our microscopy analysis was about 44âbp per min (around 1.3âkb per 30âmin).
Categorization of replicated DNA fibres and the measurement of IOD and fork speed
Images of DNA fibres were subjected to auto-thresholding with the âminimumâ or âdefaultâ algorithm in Fiji software. Individual DNA fibres were identified as a series of linearly arranged Alexa Fluor 647 (ssDNA) âdotâ signals (Fig. 2h). The thresholded fibres containing both Alexa Fluor 488 (IdU+CldU) and Alexa Fluor 594 (CldU) signals were manually categorized into those with âimmobileâ, âintermediateâ and âmobileâ class forks. The immobile class forks were defined as those with single dot signals of IdUâ+âCldU or CldU with gaps between dots, reflecting extremely slow fork movement. Here, gaps were defined as those with at least one dot of Alexa Fluor 647 ssDNA signal (Fig. 2h (immobile)). The mobile class forks were defined as those with a series of dot signals (â¥2âdots) of IdUâ+âCldU that contain a series of dot signals (â¥2âdots) of CldU on either side of the same DNA fibre (Fig. 2h (mobile)). The intermediate class forks were defined as those with an intermediate character between the two other classes; these fibres also contained dot signals of IdUâ+âCldU and CldU with gaps between dots but also contained some IdUâ+âCldU single colour signals (that is, IdU-only regions) in between these dot signals. The IOD measurement method on the mobile fork class fibres is provided in Extended Data Fig. 5b. The IOD between the immobile forks was calculated by measuring the distance between the brightest pixels in the centre of the dots using the Fiji software. To determine the fork speed of the mobile forks, CldU tracks flanked by IdU tracks were identified, their lengths were measured, and were divided by the duration of the second pulse (30âmin). CldU tracks that had no ssDNA signals ahead of them were excluded from the fork speed measurement as these fibres may have been broken in the middle of the CldU track. For the immobile fork speed measurement, the details are provided in Supplementary Note 2.
Intra-S-phase checkpoint analysis
For the experiments described in Extended Data Fig. 5dâi, MC12 cells were cultured on a glass-bottomed dish, treated with 0.5âμM nocodazole for 17âh to synchronize in prometaphase and subjected to 3âμgâmlâ1 aphidicolin treatment for 3âh. After synchronization at the G1/S-phase border, the cells were further cultured in the presence of aphidicolin for 15âh with or without 10âmM 2-aminopurine (2-AP; an intra-S-phase checkpoint inhibitor). After removing aphidicolin or 2-AP, the cells were labelled with 20âμM EdU for 60âmin and stained with EdU. The C57BL/6 strain 4-cell and 8-cell embryos within 1â2âh after the 2-to-4-cell or 4-to-8-cell division (that is, in early S phase) were treated with 3âμgâmlâ1 aphidicolin with or without 2-AP for 5.5âh followed by reagent removal and EdU staining.
Live-cell imaging
After linearization of the template plasmid, mRNA was synthesized using the mMESSAGE mMACHINE KIT (Ambion). Synthesized RNAs were stored at â80â°C until use. In vitro-transcribed mRNAs (0.9âpl of 150ângâµlâ1 mEGFP-SLX4, 0.9âpl of 150ângâµlâ1 mEGFP-PCNA, 0.9âpl of 150ângâµlâ1 Major-satellite-mClover and 0.9âpl of 35ângâµlâ1 H2B-mCherry) were microinjected into 1-cell embryos. Live-cell imaging was performed as previously described66 with some modifications. In brief, a Zeiss LSM710, LSM780 or LSM880 confocal microscope equipped with a 40à C-Apochromat 1.2NA water-immersion objective lens (Carl Zeiss) was controlled by a multi-position autofocus macro67 for Zen Software (Carl Zeiss). For major-satellite imaging, 17 confocal z sections (every 1.5âµm) of 512âÃâ512 pixel xy images covering a total volume of 30.30âÃâ30.30âÃâ24.00âµm were acquired at 2âmin 15âs intervals for at least 3âh just after nuclear envelope breakdown. For SLX4 imaging, 17 confocal z sections (every 2âµm) of 512âÃâ512âpixel xy images covering a total volume of 30.30âÃâ30.30âÃâ32.00âµm were acquired at 3âmin intervals for at least 10âh. For PCNA imaging, 29 confocal z sections (every 3âµm) of 512âÃâ512 pixel xy images covering a total volume of 84.85âÃâ84.85âÃâ84.00âµm were acquired at 5âmin intervals from the 1-cell to 16-cell stages. In Figs. 3c,g and 4j and Extended Data Fig. 7a, to achieve high-resolution live imaging while minimizing phototoxicity, we selected and imaged blastomeres (cells) that were just entering M phase and close to the objective lens, up to two blastomeres (cells) per embryo.
3D imaging analysis
To detect chromosome aberration using live imaging (Fig. 3c), we analysed images of embryos expressing major-satellite-mClover and H2B-mCherry using Imaris software (Bitplane). Chromosome bridges were detected at anaphase timepoints. To analyse DNA repair foci at M phase (Fig. 3f), we used images of embryos expressing mEGFPâSLX4 and H2BâmCherry. To detect DNA repair foci, images at 6âmin after nuclear envelope breakdown were processed using the 3D Spots detection function in Imaris software with a threshold of 2.0 times the cytoplasmic signal intensity. The detected spots were manually checked for quality and tracked over time through prometaphase until they disappeared. At each timepoint, the number of SLX4 foci on chromosomes was counted. To measure the cell cycle progression from the 1- to 8-cell stage (Fig. 4 and Extended Data Fig. 7), we analysed images of embryos expressing mEGFPâPCNA and H2BâmCherry with Imaris. To determine the timing of late S, images were processed using the â3D Spotsâ function in Imaris with a threshold of 6.0 times the cytoplasmic intensity. The detected spots were manually checked for quality. The timing when the first PCNA spot appeared was defined as the onset of late S, whereas the timing when the last spot disappeared was defined as the onset of G2. We tracked all cells in an embryo while detecting chromosome bridges at each cell division. In Fig. 4, we defined error cells as those that exhibited chromosome bridges for the first time during the 4-to-8-cell division.
scRepli-seq data analysis
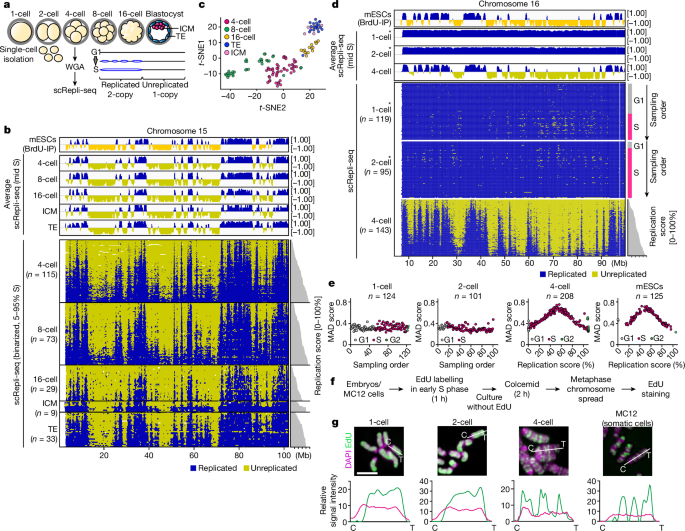
In brief, after NGS, the raw scRepli-seq FASTQ files were processed for adapter trimming of both Illumina and SEQXE adapters, mapped to the mm9 reference genome and we filtered out the duplicated reads and reads that overlapped with the mm9 blacklists as described previously16. For quality control of scRepli-seq data, we applied MAD-score-based screening to filter out cells with problematic data (MAD scores of 0 or >1.0). More than 90% of cells in each sample passed these criteria. To generate log2[median] single-cell RT profiles, we counted the reads in sliding windows of 200âkb at 40âkb intervals after normalizing S-phase data with AneuFinderâs correctMappability command based on G1 control without karyotype defects in each strain. The binarization using 80âkb or 400âkb (haplotype-resolved analysis) windows was performed using the findCNVs command in AneuFinder as described previously16. For 4-cell embryos, we applied the 1-somy mode for early-S-phase cells and the 2-somy mode for mid/late-S-phase cells. For 1- and 2-cell embryos, we used the 2-somy mode for binarization (that is, the default copy number is âreplicatedâ). As such, the overall binary profile will become blue (replicated) if there is no copy-number variation; likewise, if we use the â1-somyâ mode for the analysis of 1- and 2-cell embryos, it will be all yellow (unreplicated) instead of blue (Extended Data Fig. 2c). If we use a third colour to describe the peculiar replication regulation of the 1- and 2-cell S phase, we thought that it would be confusing. Thus, to highlight the binarization failure and the unconventional replication regulation in 1- and 2-cell-stage embryos, we decided to use the â2-somyâ mode and blue (replicated) to describe the copy-number state of the majority of bins (Figs. 1d and 2f and Extended Data Figs. 1g, 2e,h and 4a). The percentage replication scores (that is, the percentages of all of the genomic bins that have completed their replication) were calculated from binarized scRepli-seq data (excluding chromosome X) as described previously16. Averaged scRepli-seq profiles shown in Fig. 1b and Extended Data Fig. 1d were calculated from cells with 30â70% replication scores (excluding chromosome X). To identify the heterogeneously late-replicating domains described in Fig. 2f and Extended Data Figs. 1g and 2h, scRepli-seq profiles obtained from cells throughout the S phase were used to calculate the averaged scRepli-seq profile. The tag-density profile was generated in sliding windows of 200âkb at 40âkb intervals using AneuFinder as described previously16. t-SNE clustering analysis of scRepli-seq data (excluding chromosome X) was performed using RtSNE. For RtSNE, log2[median] RT scores of mid-S cells (cells with 30â70% replication scores) were used that were obtained from 4-, 8-, 16-cell embryos, ICM and TE.
Hi-C data analysis
Principal component 1 (PC1; A/B compartment profile) of Hi-C data in 200âkb bins was computed from the .hic file using published mapped Hi-C datasets of sperm68 and cumulus60 cells as described previously16. The genomic coordinates of PC1 profiles were converted from mm10 to mm9 using the liftover tool (UCSC Genome Browser). In Extended Data Figs. 2k and 4c, we defined the four PC1 (A/B compartment) categories, A1, A2, B2 and B1, as those containing 25% of all PC1 values (200âkb bins) each from the highest (strongest A) to lowest (strongest B) without an overlap.
Chromosome aberration analysis by scRepli-seq
To analyse chromosome aberration, we used the findCNVs command in AneuFinder with a 500âkb bin size as described previously16 (6-HMM options: method=âHMMâ, max.iter=3000, states=c(âzero-inflationâ, â0-somyâ, â1-somyâ, â2-somyâ, â3-somyâ, â4-somyâ, â5-somyâ, â6-somyâ), eps=0.01). Using scRepli-seq data of 4-, 8- and 16-cell-stage embryos, we determined the cell division that produced de novo chromosome errors as follows. When a pair of cells within an embryo was found to exhibit a 3:1 copy-number ratio (3-somy:1-somy) in a complementary manner for a particular chromosome or a chromosomal region, these cells were judged to be a pair of sister cells that experienced a chromosome segregation error for the first time in the last cell division (shown in Fig. 3b as de novo chromosome aberrations). When identical chromosomal abnormalities were commonly found in multiple pairs of cells within an embryo, these cells were judged to have experienced errors in divisions preceding the last division. Details of all of the detected chromosome aberrations are shown in Extended Data Fig. 6b and Supplementary Table 1. As we pooled embryos from multiple females before single-embryo sampling, our dataset is not suitable for female-to-female variability assessment.
Statistics and reproducibility
Statistical analyses were performed with GraphPad Prism v.7.02 using one-way ANOVA with Dunnâs multiple-comparison test (Figs. 2jâl, 3gâi and 4h,I); two-tailed unpaired Studentâs t-tests (Fig. 4d); two-tailed unpaired Ï2 tests (Fig. 4f); two-tailed MannâWhitney U-tests (Fig. 4g); and two-tailed Fisherâs exact tests (Fig. 4j).
Experimental reproducibility was demonstrated as follows: Fig. 1g, two independent experiments; Fig. 2d, two (2-cell) and three (1-cell, 4-cell) independent experiments; Fig. 2l, two independent experiments; Fig. 3b, two independent experiments; Fig. 3c, four independent experiments; Fig. 3g, two (2-to-4-cell and 16-to-32-cell) and three (4-to-8-cell and 8-to-16-cell) independent experiments; Fig. 3h, two independent experiments; Fig. 3i, two independent experiments; Fig. 4aâd, two independent experiments; Fig. 4h, two independent experiments; Fig. 4i, two independent experiments; and Fig. 4j, four independent experiments.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.