Overview of hifiasm (ONT)
The existing hifiasm assembly toolkit consists of three approaches: the original hifiasm6, hifiasm (Hi-C)8 and hifiasm (UL)1, each designed for specific purposes. These methods are proposed, respectively, for trio-binning haplotype-resolved assembly using parental data, single-sample haplotype-resolved assembly using Hi-C reads and T2T hybrid assembly. The core component shared by these methods is constructing a high-quality assembly graph through de novo assembly of PacBio HiFi reads. However, owing to the limited length of PacBio HiFi reads, long, repetitive genomic regions often cannot be fully resolved within this core assembly graph.
To address this limitation, longer but less accurate ONT simplex reads, especially ultra-long reads, have been used by existing hybrid T2T assemblers such as hifiasm (UL)1 and Verkko2,10. However, these assemblers do not make full use of ONT ultra-long reads because they cannot perform de novo assembly with them directly. The higher error rate and recurrent sequencing errors of ONT reads pose challenges for distinguishing genomic variations from sequencing errors. As a result, both hifiasm (UL) and Verkko use a two-stage strategy: first, constructing an assembly graph from PacBio HiFi reads with de novo assembly; and second, aligning ONT ultra-long reads to resolve regions that are unresolved by HiFi reads alone. For ONT reads, this alignment-based approach remains inherently limited by inaccuracies and biases introduced in the HiFi-based assembly graph.
Our hifiasm (ONT) approach enhances this core capability by enabling effective de novo assembly of ONT simplex reads, making full use of their longer length. Compared with HiFi-based assembly graphs, ONT-based assembly graphs tend to be more accurate and cleaner, and to resolve more repetitive regions. This improved ONT-based assembly graph substantially improves existing methods in the hifiasm toolkit, such as trio-binning or Hi-C phased assembly. The specific strategies of hifiasm (ONT) for using ONT simplex reads in T2T assembly are detailed below.
Error correction of ONT simplex reads
Error correction generates near-error-free reads by fixing sequencing errors in raw data, which is a crucial step for genome assembly. To correct a given target read R, assembly algorithms need to first collect all related reads originating from the same genomic region. Conventional algorithms assume that any overlapping read belongs to the same genomic region as R if they share high sequence similarity. However, this approach fails to distinguish highly similar repeat copies or haplotypes. As a result, many false-positive reads from other repeats or haplotypes might be incorrectly used to correct R, leading to an overcorrection issue that might collapse repeats and haplotypes in the final assembly.
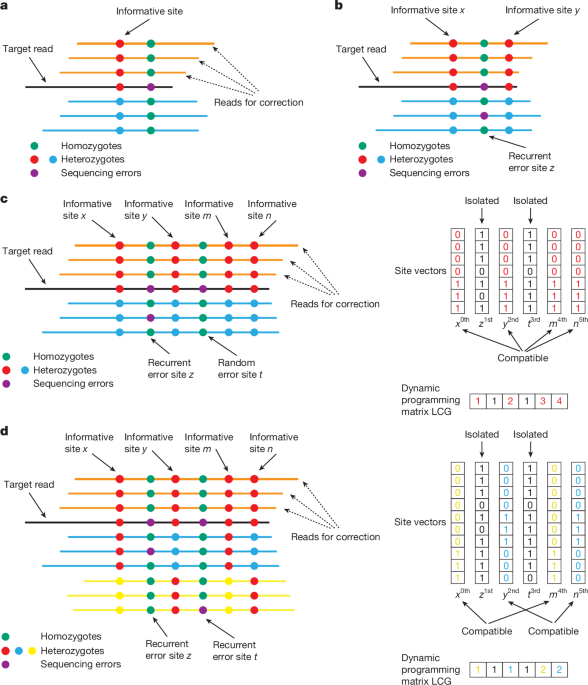
To address this problem when assembling PacBio HiFi reads, hifiasm uses a key assumption that most sequencing errors in PacBio HiFi data occur randomly and typically appear in only a single read. Figure 1a shows an example. When correcting the target read R, hifiasm identifies mismatches through pairwise alignments between R and all overlapping reads. Mismatches that are supported by multiple overlaps are considered as informative sites representing true genomic variants, whereas those that appear in only one read are treated as sequencing errors and ignored. This strategy is essentially similar to widely used variant-calling methods. Subsequently, only overlapping reads showing no differences at these informative sites compared to R are used for correction.
However, this existing hifiasm error-correction strategy is unsuitable for ONT simplex reads (Fig. 1b). Although the overall sequencing error rate of ONT simplex reads is not significantly higher than that of PacBio HiFi reads, ONT simplex reads exhibit a higher frequency of recurrent, non-random errors. Therefore, the current hifiasm method would mistakenly identify these recurrent errors as informative sites. For a target simplex read R, overlapping reads originating from the same genomic region would thus be incorrectly discarded if they differed from R at these recurrent error sites. As a result, most ONT simplex reads would remain uncorrected, and could not be used for de novo genome assembly.
In hifiasm (ONT), we introduce an approach that makes use of the long-range phasing information of ONT reads to improve error correction. The basic idea is that true informative sites representing real genomic variants usually appear together and are mutually compatible with other informative sites. In practice, hifiasm (ONT) clusters potential informative sites on the basis of their compatibility. As shown in Fig. 1c, given a target ONT simplex read awaiting correction, overlapping reads at a potential informative site (x, y, m, n, z or t) can be classified into two phases: phase 0 (matching the target read) and phase 1 (differing from the target read). For two sites, if both consistently classify overlapping reads into identical phases, they are considered compatible and grouped together. True variants are expected to be compatible with multiple other real variant sites. By contrast, isolated sites that lack compatibility with others (such as z and t) have a higher likelihood of representing sequencing errors. This method is conceptually similar to using haplotype phasing to improve the accuracy of variant calling. To further enhance reliability, hifiasm (ONT) adopts the following criteria: (i) an isolated site is considered informative only if it is supported by a sufficiently high number of reads; and (ii) any grouped site that is supported by more than one read is regarded as an informative site, because such sites are inherently more reliable.
In practice, it is necessary to develop an efficient algorithm for grouping potential informative sites, because this operation must be performed for each read during error correction. To achieve this, we propose a dynamic programming method designed to identify the largest compatible group for each site. Specifically, let R be the target read awaiting correction, and S be the list of N potential informative sites within R, sorted by their positions. Here, S[i] denotes the i-th site in the list, and S[i][k] represents the phase assignment of the k-th read at site S[i]. The value of S[i][k] can take one of the following states:
$$S[i][k]\in \{0,1,* \},$$
where 0 and 1 indicate that the k-th read is assigned to phase 0 or phase 1, respectively, and * indicates that the k-th read does not cover site S[i]. The details of the dynamic programming method are described as follows:
-
1.
Subproblem. Let LCG[i] be the size of the largest compatible group in S that ends at index i and is compatible with S[i]. The goal is to compute LCG[0] to LCG[N − 1] for all sites and identify those with values greater than 1, which indicate a compatible group rather than an isolated site.
-
2.
Recurrence relation. Formally, the recurrence relation is defined as follows:
$$\mathrm{LCG}(i)=\mathop{\max }\limits_{\genfrac{}{}{0ex}{}{j < i}{S[\,j]\leftrightarrow S[i]}}\{\mathrm{LCG}(j)\}+1,$$
where \(S[\,j]\leftrightarrow S[i]\) indicates that S[j] is compatible with S[i]. Two sites S[j] and S[i] are considered compatible if and only if
$$S[i][k]=S[\,j][k]\,\mathrm{for}\,\mathrm{all}\,k,\mathrm{such}\,\mathrm{that}\,S[i][k]\in \{0,1\}\,\mathrm{and}\,S[j][k]\in \{0,1\}.$$
Figure 1c provides an example of the dynamic programming matrix LCG.
-
3.
Traceback for grouping sites. Hifiasm (ONT) identifies any entry where LCG[i] > 1, starting from the highest value and proceeding downward. For each site S[i] with LCG[i] > 1 that has not yet been assigned to a cluster, the algorithm traces back through its compatible prefix sites S[j], following the path used to compute LCG(i). For example, in Fig. 1c, hifiasm (ONT) starts from LCG(5), which holds the highest score, and groups the corresponding sites S[5], S[4], S[2] and S[0] (that is, n, m, y and x) during the traceback process.
The time and space complexity of this dynamic programming method are O(n2) and O(n), respectively, making it efficient for error correction in de novo assembly. An additional advantage of this approach is that it does not rely on the diploid-genome assumption, enabling it to handle polyploid genomes or highly similar repeats with more than two repeat copies. As shown in Fig. 1d, hifiasm (ONT) successfully identifies x and m as one group and n and y as another group when there are three haplotypes available.
We further improve the error correction by filtering out low-quality base pairs as follows:
-
1.
Potential homopolymer sequencing errors. ONT reads are known to exhibit a high sequencing error rate within homopolymer regions. If a potential informative site is located in a homopolymer region, hifiasm (ONT) discards it, because it is more likely to result from homopolymer-induced sequencing errors.
-
2.
Strand bias. Given the target read R, hifiasm (ONT) excludes an informative site if all reads that support R originate from one strand, whereas all other reads that differ from R at this site originate from the opposite strand. Strand bias is a common sequencing error observed in ONT reads.
-
3.
Low base quality score. In addition to sequence data, hifiasm (ONT) also loads base quality scores into memory. Any base pair with a quality score lower than 10 is considered a potential sequencing error and excluded from the calculation of informative sites.
One potential challenge for hifiasm (ONT) arises when sequencing errors occur at the exact position of a true variant, making that variant incompatible with others. However, in practice, such cases are rare. Even if a true variant in one read is affected by sequencing errors, other nearby variants that remain unaffected can still be accurately detected, allowing hifiasm (ONT) to effectively separate haplotypes and resolve repeat copies.
Improved strategies for T2T assembly
With the error-correction approach in hifiasm (ONT), most sequencing errors within ONT simplex reads can be corrected effectively. These nearly error-free reads are then used with the existing assembly strategies in hifiasm to construct a high-quality assembly graph. For haploid genomes, a graph cleaning strategy is applied to produce linear assembly results. For diploid genomes, further data—such as parental or Hi-C reads—are required to generate haplotype-resolved assemblies using hifiasm’s existing trio-binning or Hi-C phasing approaches.
To further improve T2T assembly, we developed a strategy to retain telomere sequences in the final assembly. A common issue in hifiasm is that although it can reconstruct entire chromosomes, it can still miss telomeric sequences at chromosome ends. This occurs because in the assembly graph—particularly for diploid genomes—telomere ends often appear as tips. During graph cleaning, hifiasm typically discards these tips, because most are caused by assembly errors. As a result, telomere sequences might be inadvertently removed from the final assembly. To address this, the improved T2T assembly strategy in hifiasm (ONT) first checks whether any reads contain telomeric sequences before the assembly. If such reads are detected, hifiasm preserves the corresponding graph tips during graph cleaning. This approach helps to retain more telomere ends and results in an increased number of T2T contigs and scaffolds.
We also developed a dual-scaffold approach to assemble more chromosomes from telomere to telomere at the scaffold level. The goal is to scaffold gapless contigs into longer, gapped scaffolds by using information from both haplotypes. The basic idea is that, for an assembly gap in haplotype 1, the dual-scaffold approach examines the corresponding homologous regions in haplotype 2. If the region in haplotype 2 is completely assembled without gaps, the dual-scaffold method fills the gap in haplotype 1 with ambiguous nucleotides (Ns), using the estimated length inferred from the complete sequence in haplotype 2. In essence, this approach performs reference-guided scaffolding for each haplotype41, using the other haplotype as a reference.
ONT sequencing and basecalling
ONT standard simplex sequencing data for the GIAB samples HG001–HG007 have been deposited in the official ONT open data repository (s3://ont-open-data/giab_2025.01/). Cell lines for these samples were obtained from the Human Genetic Cell Repository at the Coriell Institute for Medical Research and cultured according to the supplier’s recommended protocols. High-molecular-weight DNA was extracted using the QIAGEN Puregene cell extraction kit, followed by library preparation with the SQK-LSK114 kit according to ONT protocols, and sequencing was performed on PromethION flow cells using P48 instruments. Basecalling was done using Dorado v.0.7.2 with both HAC v.5.0.0 and SUP v.5.0.0 models. For HG001, HG003, HG004, HG005, HG006 and HG007, reads from two flow cells were basecalled and used for assembly. For HG002, only data from a single flow cell were used, because one flow cell was sufficient to produce ONT simplex reads at approximately 50× coverage. In addition, we re-basecalled the existing D. rerio (zebrafish) dataset using Dorado v.0.8.3 with the SUP v.5.0.0 model to improve read-level base accuracy.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.