All consumables, such as antibodies, chemicals and so on can, be found in the resource list in Supplementary Table 6.
Animal experiments
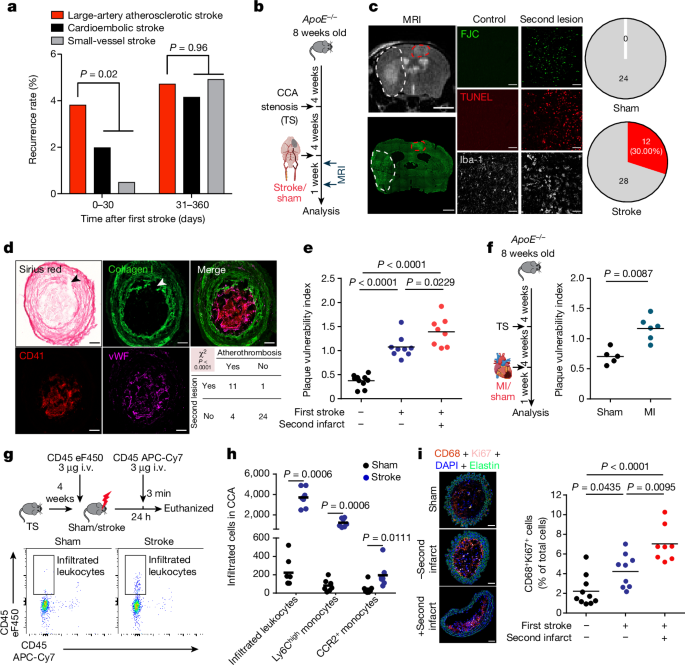
All animal experiments were performed in accordance with the guidelines for the use of experimental animals and were approved by the government committee of Upper Bavaria (Regierungspraesidium Oberbayern; #02-2018-12). All mice used in this study were between 6 and 16 weeks of age independent of genotype and experimental setup. ApoEâ/â (B6.129P2-Apoetm1Unc/J; JAX strain: 002052), WT (C57BL/6J; JAX strain: 000664), Aim2â/â (B6.129P2-Aim2Gt(CSG445)Byg/J; JAX strain: 013144), Pycardâ/â (Ascâ/â; B6.129S5-Pycardtm1Vmd) and R26-CAG-ASC-citrine (B6.Cg-Gt(ROSA)26ortm1.1(CAG-Pycard/mCitrine*,-CD2*); JAX strain: 030744) mice were bred and housed at the animal core facility of the Centre for Stroke and Dementia Research (Munich, Germany). Ldlrâ/â:Mx1cre:c-Mybfl/fl mice were bred and housed at the animal facility of Walter Brendel Centre (Munich, Germany). ApoEâ/â mice were fed an HFD (#88137, ssniff) from 8 weeks onwards. cGasâ/â (B6(C)-Cgastm1d(EUCOMM)Hmgu/J), Nlrp1â/â (Del(11Nlrp1a-Nlrp1c-ps)1Smas) and Nlrp3â/â (C57BL/6J-NLRP3tm1Tsc) mice were bred and housed at the Gene Centre of the LMU University Munich (Germany).
For this exploratory study, animal numbers were estimated based on previous results from the transient ischaemiaâreperfusion stroke model on extent and variability of atheroprogression after stroke. Data were excluded from all mice that died during surgery. Detailed exclusion criteria are described below. Animals were randomly assigned to treatment groups, and all analyses were performed by investigators blinded to group allocation. All animal experiments were performed and reported in accordance with the Animal Research: Reporting of In Vivo Experiments (ARRIVE) guidelines35.
Drug administrations
Oral gavage with aspirin and rosuvasatin
Mice received a daily bolus of aspirin (20âmgâkgâ1, Sigma Aldrich) and rosuvastatin (5âmgâkgâ1, Sigma-Aldrich) via oral gavage. Aspirin and rosuvastatin were solved in water (sterile ddH2O) and mixed with a powdered chow diet (ssniff). A single daily bolus was 500âµl.
Recombinant human DNase I
ApoEâ/â mice received DNase injections as we previously described18. In brief, 1,000âU recombinant DNase I (Roche) dissolved in incubation 1à buffer (40âmM Tris-HCl, 10âmM NaCl, 6âmM MgCl2 and 1âmM CaCl2, pH 7.9, diluted in PBS; Roche) was injected intravenously via tail vein right before surgery in a final volume of 100âμl. The control group was administered saline injections at the same volume, routine and timing as the experimental group.
Caspase-1 inhibitor (VX765)
The caspase-1 inhibitor VX765 (stock in DMSO) was dissolved in PBS (Belnacasan, Invivogen) and injected intraperitoneally 1âh before surgery at a dose of 100âmgâkgâ1 body weight at a final volume of 300âµl (ref. 18). The control group was administrated saline injections at the same volume, routine and timing as the experimental group.
NLRP3 inflammasome inhibitor (MCC950)
Mice received two injections (1âh before and 1âh after surgery) of the NLRP3 inflammasome inhibitor MCC950 dissolved in sterile saline at a dose of 50âmgâkgâ1 body weight (Invivogen). MCC950 or the control (sterile saline) was injected intraperitoneally in a final volume of 200âµl.
AIM2 inhibitor (4-sulfonic calixarene)
The AIM2 inhibitor 4-sulfonic calixarene has recently been characterized by Green et al.36. The stock solutions (in DMSO) were dissolved in PBS and injected intraperitoneally 1âh before surgery at a dose of 5âmgâkgâ1 body weight at a final volume of 200âµl. The control group was administered control injections (sterile saline) at the same volume, routine and timing as for the intervention group.
DNA challenge
DNA derived from stimulated neutrophils (see below for neutrophil stimulation and isolation of NETâDNA) was injected intravenously at a dose of 5âµg at a final volume of 200âµl (sterile saline). The control group was administrated control injections (sterile saline) at the same volume, routine and timing as for the intervention group.
Neutrophil depletion
Neutrophils were depleted using a specific antibody to Ly6G (clone: 1A8, BioXCell). Mice were administered at a concentration of 14âmgâkgâ1 body weight at a final volume of 250âµl intraperitoneally. The control group was administrated control IgG injections at the same volume, routine and timing as the experimental group.
PAD4 inhibitor (GSK484)
The PAD4 inhibitor GSK484 (stock in 100% EtOH) was diluted in PBS to a final concentration of 4âmgâkgâ1 body weight at a final volume of 100âµl. GSK484 was administered 1âh before surgery. For more than 6âh of survival time, a second bolus (4âmgâkgâ1 body weight) was injected intraperitoneally 4âh post-surgery. The control group was administrated control injections (sterile saline) at the same volume, routine and timing.
IL-1β neutralization
Mice received two injections of antagonizing anti-IL-1β in sterile saline (clone: B122, BioXCell), 1âh before and 1âh after surgery. Anti-IL-1β or the corresponding IgG control was injected intraperitoneally at a dose of 4âmgâkgâ1 body weight in a final volume of 200âµl.
Patient cohorts for epidemiological analysis
The analysis presented in Fig. 1a was performed using patient data from two multicentre prospective observational hospital-based cohort studies of patients with acute ischaemic stroke in Germany (PROSCIS and DEMDAS/DEDEMAS). All patients provided written informed consent. Patient characteristics are provided in Supplementary Table 1. The study protocols and the detailed baseline patient characteristics have been previously described13,14. Basic demographic and stroke-related characteristics are summarized below. In brief, for both the PROSCIS and the DEMDAS/DEDEMAS cohorts, patients 18 years of age or older with an acute ischaemic stroke confirmed with neuroimaging and with symptom onset in the past 7 and 5 days, respectively, were recruited through the local stroke units of seven tertiary stroke centres in Germany. Patients in PROSCIS underwent follow-up telephone interviews at 3 and 12 months after stroke, whereas patients in DEMDAS/DEDEMAS underwent a telephone interview at 3 months after stroke as well as face-to-face interviews and inspection of their medical records by a physician at 6 and 12 months after stroke. The outcome of interest for the current analysis included the occurrence of a recurrent ischaemic stroke or transient ischaemic attack within the first 12 months after stroke, as self-reported by the patient and documented in their medical records.
Patient cohort for carotid endarterectomy sample analysis
Patients scheduled for carotid endarterectomy due to symptomatic or asymptomatic carotid stenosis were prospectively recruited at the Department of Neurology and Cardiothoracic Transplantation and Vascular Surgery at Hannover Medical School between June 2018 and December 2020. Patients characteristics are provided in Supplementary Table 2. Carotid stenosis was defined as symptomatic if cerebral ischaemia occurred in the territory of the affected artery, and concurrent stroke aetiologies were excluded following standardized stroke diagnostics including cranial computed tomography and/or MRI, computed tomography or MR-angiography, transthoracic or transoesophageal echocardiography, cardiac rhythm monitoring and Doppler/duplex ultrasound. Peripheral venous blood was drawn immediately before surgery, and EDTA whole-blood samples were used for flow cytometry analysis. Carotid plaque samples were obtained during carotid endarterectomy and immediately preserved in PBS. Both blood and tissue samples were sent for further analysis on the same day of collection. All patients provided written informed consent and the ethics committee at Hannover Medical School approved the study.
Thirteen patients with symptomatic and seven patients with asymptomatic, high-grade carotid stenosis were recruited. The median age was 73 years (25thâ75th percentile: 62â80 years of age). See Supplementary Table 2 for clinical and demographic patient details.
Patient cohort for myocardial infarction sample analysis
Patients with ST-elevation myocardial infarction (STEMI) were prospectively recruited between September 2016 and February 2018 at the German Heart Centre Munich and the Klinikum rechts der Isar (both at the Technical University of Munich). Patients characteristics are provided in Supplementary Table 3.
The diagnosis of STEMI was based on chest pain within the past 12âh, persistent STâsegment elevation of 1âmm or more in at least two extremities or 2âmm or more in at least two chest leads and diagnosis of type 1 myocardial infarction according to cardiac catheterization. Exclusion criteria were: cardiogenic shock, left ventricle ejection fraction of 35 or less, coexisting chronic or inflammatory diseases, anti-inflammatory drug therapy (for example, cortisol) and myocardial infarction type 2â5. Blood samples for plasma analysis were collected in EDTA tubes immediately after admission to the hospital or latest 6âh after coronary intervention.
Age-matched and sex-matched patients with known stable coronary artery disease served as controls. They were prospectively recruited between February 2017 and February 2018 during consultation in the outpatient department of the German Heart Centre Munich for routine examination. Exclusion criteria were: history of myocardial infarction, reduced left ventricle ejection fraction, chronic or inflammatory diseases, and anti-inflammatory drug therapy. Blood samples for plasma analysis were collected in EDTA tubes on the day of consultation in the outpatient department. All patients provided written informed consent and the institutional ethics committee at Technical University Munich approved the study (235/16S). EDTA tubes of both STEMI and control patients were centrifuged at 4â°C and 1,600g for 30âmin directly after collection. Plasma aliquots were stored at â80â°C for further analysis.
Carotid tandem stenosis model
Tandem stenosis surgery was performed as previously described37. In brief, at 12 weeks of age, 4 weeks after commencement of an HFD, ApoEâ/â mice (C57BL/6J background) were anaesthetized with 2% isoflurane delivered in a mixture of 30% and 70% N2O. An incision was made in the neck, and the right common carotid artery was dissected from circumferential connective tissues. To control the stenosis diameter, a 150-µm or 450-µm dummy was placed on top of the exposed right common carotid artery, with the distal point 1âmm away from the bifurcation and proximal point 3âmm from the distal stenosis; subsequently, a 6-0-braided polyester fibre suture was tied around both the artery and the needle, and then the pin was carefully removed. Animals were fed with an HFD for an additional 4 weeks after tandem stenosis surgery.
Bone marrow transplantation
Donor animals (B6.129P2-Aim2Gt(CSG445)Byg/J or C57BL/6J) were euthanized and the femur, tibia and humerus were collected in cold PBS. Bone marrows were isolated from bones and filtered through 40-μm cell strainers to obtain single-cell suspensions. After washing, cell number and viability were assessed using an automated cell counter (Countess 3, Thermo Fisher Scientific) and Trypan Blue solution (Merck). Cells were injected intravenously into Mx1cre:c-Mybfl/fl recipient mice (Ldlrâ/â background; 8â15âÃâ106 cells per mouse) in a total volume of 100âµl saline. At the time of transplantation, recipient mice had previously been treated with poly(I:C) solution at a dose of 10âμgâgâ1 body weight every other day five times to induce bone marrow depletion38. Mice were fed with a HFD and maintained for 4 weeks after transplantation to establish efficient bone marrow repopulation.
Ischaemiaâreperfusion stroke model
Four weeks after tandem stenosis surgery, ApoEâ/â mice were anaesthetized with 2% isoflurane delivered in a mixture of 30% O2 and 70% N2O. An incision was made between the ear and the eye to expose the temporal bone. Mice were placed in supine position, and a laser Doppler probe was attached to the skull above the middle cerebral artery (MCA) territory. The common carotid artery and left external carotid artery were exposed via middle incision and further isolated and ligated. A 2-mm silicon-coated filament (Doccol) was inserted into the internal carotid artery, advanced gently to the MCA until resistance was felt, and occlusion was confirmed by a corresponding decrease in blood flow (that is, a decrease in the laser Doppler flow signal by 80% or more). After 60âmin of occlusion, the animals were re-anaesthetized and the filament was removed. After recovery, the mice were kept in their home cage with ad libitum access to water and food. Sham-operated mice received the same surgical procedure, but the filament was removed in lieu of being advanced to the MCA. Body temperature was maintained at 37â°C throughout surgery in all mice via feedback-controlled heating pad. Exclusion criteria included: (1) insufficient MCA occlusion (a reduction in blood flow to more than 20% of the baseline value), (2) death during the surgery, and (3) lack of brain ischaemia as quantified post-mortem by histological analysis.
Myocardial infarction
Myocardial infarction surgery was performed as previously described39. In brief, mice were intubated under MMF anaesthesia (midazolam 5.0âmgâkgâ1 body weight, medetomidine hydrochloride 1.0âmgâkgâ1 body weight and fentanyl citrate 0.05âmgâkgâ1 body weight; intraperitoneally) and thoracotomy was performed in the left intercostal space. The left anterior descending coronary artery was identified and myocardial infarction was induced by permanent ligation with an 8-0 prolene suture. Atipamezole hydrochloride (5âmgâkgâ1 body weight) and flumazenil (0.1âmgâkgâ1 body weight) injected subcutaneously were used to antagonize MMF anaesthesia. Mice received subcutaneous buprenorphine (0.3âmgâkgâ1 body weight) as an analgesic every 8âh for 3 days starting at the end of the surgical procedure.
Ultrasound imaging (mouse carotid artery Doppler analysis)
Carotid artery blood flow in ApoEâ/â mice was measured with a high-frequency ultrasound imaging system (Vevo 3100LT, Visual Sonics) with a 40-MHz linear array transducer (MX550D, Visual Sonics) before and right after tandem stenosis surgery, and weekly for 4 weeks afterwards. Mice were anaesthetized with isoflurane delivered in a mixture of 30% O2 and 70% N2O. B-mode, colour-Doppler mode and pulsed Doppler velocity spectrum were recorded from both sides of the CCA. For the right CCA (RCCA), five locations were examined: before proximal ligation, near proximal ligation, between two ligations, near distal ligation and above the distal ligations. For the left CCA (LCCA), as it was not ligated, only three locations were measured: proximal, middle and distal part of the LCCA. Pulsed Doppler velocity was determined with the sample volume calibrated to cover the entire vascular lumen and the smallest possible angle of interception (less than 60°) between the flow direction and the ultrasound beam. The peak systolic velocity (PSV) and end diastolic velocity (EDV) were recorded from CCAs of both sides. VevoLab v3.2.0 software was used for ultrasound imaging analysis. The mean velocity was calculated as: mean velocityâ=â(PSVâ+â2âà EDV)/3.
MRI for secondary lesion detection
MRI was performed in a small-animal scanner (3T nanoScan PET/MR, Mediso, with 25-mm internal diameter quadrature mouse head coil) at 2 and 7 days after sham or stroke surgery. For scanning, mice were anaesthetized with 1.2% isoflurane in 30% O2//70% N2O applied via face mask. Respiratory rate and body temperature (37â±â0.5â°C) were continuously monitored via an abdominal pressure-sensitive pad and rectal probe, and anaesthesia was adjusted to keep them in a physiological range. The following sequences were obtained: coronal T2-weighted imaging (2D fast-spin echo (FSE), repetition time/echo time (TR/TE)â=â3,000/57.1âms, averages 14, resolution 167âÃâ100âÃâ500âµm3), coronal T1-weighted imaging (2D FSE, TR/TEâ=â610/28.6âms, averages 14, resolution 167âÃâ100âÃâ500âµm3) and diffusion-weighted imaging (2D spin echo, TR/TEâ=â1,439/50âms, averages 4, resolution 167âÃâ100âÃâ700âµm3). MRI images were then post-processed using Image J.
Microparticle of iron oxide MRI for thrombus detection
MRI was performed as previously described24. In brief, mice were anaesthetized using isoflurane in a mixture of O2/N2O (30/70) and kept under anaesthesia during all the procedure, while maintaining a body temperature of 37â°C. Before MRI, mice were subjected to caudal vein catheterization for DNA and microsized matrix-based magnetic particle (M3P) administration. MRI was performed with a BioSpec 7T TEP-MRI system and a surface coil (Brukery). Imaging data were obtained using a TOF sequence to visualize vascular structures, a T2*-weighted sequence for iron-sensitive imaging and a T2-weighted sequence for tissue contrast. The MRI parameters were set at TR/TEâ=â12/4.2âms for the TOF sequence, TR/TEâ=â50/8.6âms for the T2*-weighted sequence and TR/TEâ=â3,500/40âms for the T2-weighted sequence. T2*-weighted images are presented as a stack of four slices (minimum intensity) after bias fields correction using ImageJ software.
Organ and tissue processing
Mice were deeply anaesthetized with ketamine (120âmgâkgâ1) and xylazine (16âmgâkgâ1) and venous blood was drawn via cardiac puncture of the right ventricle in 50âmM EDTA (Sigma-Aldrich); the plasma was isolated by centrifugation at 3,000g for 15âmin and stored at â80â°C until further use. Immediately following cardiac puncture, mice were transcardially perfused with ice-cold saline. Subsequently, the CCAs from both sides as well as the aortic arches and hearts were carefully isolated and embedded in compound (OCT, Tissue-tek), frozen over dry ice and stored at â80â°C until sectioning.
Heads were cut just above the shoulders. Skin was removed from the head and the muscle was stripped from the bone. After removal of the mandibles and the skull rostral to maxillae, the whole brain with skull was post-fixed by 4% paraformaldehyde (PFA) overnight at 4â°C. Subsequently, samples were transferred to a decalcification solution of 0.3âM EDTA (C. Roth, 292.94âgâmolâ1) at pH 7.4 and stored at 4â°C. EDTA solution was changed after 3 days. Samples were immersed with 30% sucrose in PBS and then frozen down in â20â°C isopentane (Sigma-Aldrich). Coronal sections (15âµm thick) were obtained at the level of the anterior commissure for immunohistochemical analysis. Sections were mounted on SuperfrostPlus Slides (Thermo Scientific) and stored in â80â°C.
Preparation of plasma samples for free nucleotide quantification
Mouse venous blood from cardiac puncture s drawn in 50âmM EDTA tubes. Afterwards samples were centrifuged at 1,500g for 10âmin at room temperature. Plasma was isolated, transferred to a new tube and spined again at 3,000g for 10âmin. Plasma was then carefully collected and immediately frozen down at â80â°C until further processing.
Histology and immunofluorescence
Carotid (5âµm) cryosections were histologically stained with haematoxylin and eosin (H&E) in 100-µm intervals. Total collagen content was assessed by Picro Sirius red staining (Abcam) in consecutive sections. For immunofluorescence staining, cryosections were fixed with 4% PFA followed by antigen blockade using 2% goat serum-blocking buffer containing 1% BSA (Sigma), 0.1% cold fish skin gelatin (Sigma-Aldrich), 0.1% Triton X-100 (Sigma) and 0.05% Tween-20 (Sigma). Next, sections were incubated overnight at 4â°C with the following primary antibodies: rat anti-mouse CD68 (1:200; ab53444, Abcam), mouse anti-mouse smooth muscle actin (1:200; ab7817, Abcam), rabbit anti-mouse Ki67 (1:200; 9129S, Cell Signaling), mouse anti-mouse caspase-1 (1:200; AG-20B-0042-C100, Adipogen), sheep anti-von Willebrand factor (1:100; ab11713, Abcam), rat anti-CD31 (1:200; BM4086, OriGene) and anti-collagen I (1:250; ab279711, Abcam). After washing, sections were incubated with secondary antibodies as following: AF647 goat anti-rat (1:200; Invitrogen), Cy3 goat anti-mouse IgG H&L (1:200; Abcam), AF594 goat anti-rabbit (1:200; Invitrogen), AF488 goat anti-mouse (1:200; Invitrogen) and AF594 donkey anti-sheep (1:200; Invitrogen). Counterstain to visualize nuclei was performed by incubating with DAPI (1:5,000; Invitrogen). Finally, sections were mounted with fluoromount medium (Sigma-Aldrich). Microphotographs of immunofluorescent samples were taken with a confocal microscope (LSM 880 and LSM 980; Carl Zeiss) using ZEN2 software (blue edition). Histological sections were imaged with an epifluorescent microscope (Axio Imager M2, Carl Zeiss) and quantified by using ImageJ software (US National Institutes of Health).
For the visualization of suspected secondary infarct lesions in the contralateral hemisphere, brain sections (15âµm) were first stained for Fluoro Jade C (FJC) to identify degenerating neurons. FJC staining was performed using the Fluoro-Jade C Ready-to-Dilute staining kit (TR-100-FJ, Biosensis) according to the manufacturerâs instructions. To confirm the secondary lesion, double staining of the microglia marker Iba-1 (1:200; FUJIFILM Wako Pure Chemical Corporation) and terminal deoxynucleotidyl transferase dUTP nick end labelling (TUNEL) staining was performed using the Click-iT Plus TUNEL Assay for In Situ Apoptosis Detection (Alexa Fluor 647 dye, C10619, Thermo Scientific) according to the manufacturerâs instructions. Brain samples were photographed on an epifluorescence microscope (Zeiss Axiovert 200M, Carl Zeiss) and a confocal microscope (LSM880, Carl Zeiss).
Mouse CCA plaque analysis
Plaque vulnerability was assessed as previously described15. In brief, intima, media and necrotic core area were analysed in H&E-stained sections. The necrotic core was defined as the area devoid of nuclei underneath a formed fibrous cap. Collagen content was quantified on Picro Sirius red-stained sections. The vulnerability plaque index (VPI) was calculated as VPIâ=â(% necrotic core areaâ+â% CD68 area)/(% smooth muscle actin areaâ+â% collagen area).
Flow cytometry analysis
Isolated CCA samples were mixed with digestion buffer, consisting of collagenase type XI (125âUâmlâ1, C7657), hyaluronidase type 1-s (60âUâmlâ1, H3506), DNase I (60âUâmlâ1, D5319), collagenase type I (450âUâmlâ1, C0130; all enzymes from Sigma-Aldrich) in 1à PBS40, and were digested at 750ârpm for 30âmin at 37â°C. After digestion, CCA materials were homogenized through a 40-µm cell strainer, washed at 500g for 7âmin at 4â°C and resuspended in flow cytometry staining buffer (00-4222-26, Thermo Scientific) to generate single-cell suspensions. Cell suspensions were incubated with flow cytometry antibodies and analysed using a spectral flow cytometer (Northern Light, Cytek). Alternatively, cell suspension was incubated with the fluorescent inhibitor probe 660-YVAD-FMK (#9122, Immunochemistry Technology) to label active caspase-1 in living cells. A detailed antibody list for flow cytometry analysis is available in the resource table in Supplementary Table 6.
For neutrophil flow cytometry, CCA single-cell suspensions (see above) or full EDTA blood was incubated with CD45-specific, CD11b-specific, Ly6C-specific, Ly6G-specific and citrullinated histone3 (citH3)-specific antibodies and analysed using spectral flow cytometry. Neutrophils were defined as CD45+CD11b+Ly6Ghigh cells; neutrophil activation was defined via citH3 detection.
Representative gating strategies for individual flow cytometry experiments are provided in Supplementary Fig. 2.
Analysis of plaque-infiltrating leukocytes
Circulating leukocytes were discriminated by intravenous administration of an anti-CD45 antibody41 (eFluor450, clone: 30-F11, eBioscience) immediately before stroke surgery. Twenty-four hours after stroke, to exclude the blood contamination in the CCA, an anti-CD45 antibody (APCâCy7, clone: 30-F11, BioLegend) was injected intravenously 3âmin before euthanasia. Then, CD45 eFluor450-positive but APCâCy7-negative population were considered as the âinfiltrating leukocytesâ population.
Immunoblot analysis
Ipsilateral and contralateral CCA materials were carefully isolated and snap frozen on dry ice. Whole frozen CCA was lysed with RIPA lysis/extraction buffer with added protease/phosphatase inhibitor (Thermo Fisher Scientific). Total protein was quantified using the Pierce BCA protein assay kit (Thermo Fisher Scientific). Whole-tissue lysates were fractionated by SDSâPAGE and transferred onto a polyvinylidene difluoride membrane (Bio-Rad). After blocking for 1âh in TBS-T (TBS with 0.1% Tween-20, pH 8.0) containing 4% skin mile powder (Sigma), the membrane was washed with TBS-T and incubated with the primary antibodies to the following antibodies: mouse anti-caspase-1 (1:1,000; Adipogen), rabbit anti-actin (1:1,000; Sigma) and rabbit anti-factor XII (1:1,000, Invitrogen). Membranes were washed three times with TBS-T and incubated for 1âh with horseradish peroxidase-conjugated anti-rabbit or anti-mouse secondary antibodies (1:5,000, Dako) at room temperature. Membranes were washed three times with TBS-T, developed using ECL substrate (Millipore) and acquired via the Vilber Fusion Fx7 imaging system.
ASC oligomerization assay
ASC oligomerization was performed as previously published42. In detail, bone marrow was isolated from mice and cultured for 7 days with L929 cell-conditioned medium. After differentiation, cells were washed in PBS and primed with 100ângâmlâ1 of lipopolysaccharide (LPS) for 4âh. Cells were either left primed or additionally activated by stimulation with 250âµgâmlâ1 NETâDNA for 2âh. Cells were washed in PBS and detached by scraping in PBS containing 2âmM EDTA. After centrifugation, cell pellets were resuspended in 0.5âml of ice-cold buffer A and lysed through sonification. After centrifugation to remove bulk nuclei, 20âµl of lysate was stored as input before oligomerization. Buffer A was added to the remaining lysate; following centrifugation, supernatants were diluted with CHAPS buffer and pelleted through further centrifugation. Proteins were crosslinked for 30âmin at room temperature with 50âµl of CHAPS buffer containing 4âmM disuccinimidyl suberate. After centrifugation pellets were resuspended in Laemmli buffer and boiled at 70â°C for 10âmin. Samples were loaded onto SDSâPAGE and separated at 150âV. Transfer was performed at 4â°C and 100âV for 1âh and membranes were blocked with 4% BSA in TBS-T. Membranes were incubated overnight at 4â°C with anti-ASC antibody (AL177, Adipogen), washed three times in TBS-T and incubated at room temperature for 1âh with goat anti-Rb horseradish peroxidase antibody. After washing and detecting the images, membranes were re-incubated with actinâantibody for 1âh at room temperature, proceeded by washing, secondary antibody and imaging.
ELISA
CCA tissue samples were carefully isolated and snap-frozen on dry ice. Whole frozen CCA was lysed with cell lysis buffer (#895347, R&D System). Then, the concentration of IL-1β in total CCA lysates was measured by ELISA according to the manufacturerâs instructions (MLB00C, R&D system). Absorbance at 450ânm was measured by an iMark Microplate reader (Bio-Rad).
Gelatin zymography of mouse CCA extracts, BMDMs culture medium and patient plaque lysates
CCA tissue extracts were analysed using gelatin zymography (Novex TM 10% zymography plus protein; ZY00100BOX, Thermo Scientific) according to the manufacturerâs instructions. CCA tissue was lysed with cell lysis buffer (#895347, R&D System). Total protein was quantified using a Pierce BCA protein assay kit (Thermo Fisher Scientific). Aliquots of appropriately diluted tissue extracts were loaded on gels at a total volume of 20âµl. After electrophoresis, gels were incubated in 1à Zymogram renaturing buffer (LC2670, Invitrogen) for 30âmin at room temperature with gentle agitation. Afterwards, Zymogram renaturing buffer was decanted and 1à Zymogram developing buffer (LC2671, Invitrogen) was added to the gel. The gel was then equilibrated for 30âmin at room temperature with gentle agitation. After an additional wash with 1à Zymogram developing buffer, the gels were incubated at 37â°C overnight. Gels were stained with a colloidal blue staining kit (LC6025, Invitrogen) and acquired on a gel scanner. BMDM culture medium was collected and loaded on gelatin zymography gels at a total volume of 25âµl. MMP activity in BMDM culture medium was analysed using the same protocol as for the tissue samples.
MMP2 and MMP9 in situ zymography
MMP2 and MMP9 in situ zymography on CCA sections was performed as previously described with slight modifications8. DQ-gelatin (D12054, Invitrogen) was dissolved in reaction buffer (50âmM Tris-HCl, 150âmM NaCl, 5âmM CaCl2 and 200âmM sodium azide, pH 7.6). Cryosections (5âµm) were incubated for 2âh at 37â°C with the gelatin-containing reaction buffer. Negative control sections were pre-incubated for 1âh with the MMP inhibitor 1,10-phenatheroline (Sigma). Nuclei were stained with DAPI. MMP activity was detected with an Axio Observer Z1 microscope with Ã20 magnification (Carl Zeiss). Data are shown as normalized MMP intensity (normalized MMP areaâ=âMMP+ area/intima area).
Neutrophil isolation and stimulation
Neutrophils were generated from the tibia and femur of transcardially perfused WT mice. After isolation and dissection of the tibia and femur, bone marrow was flushed out of the bones through a 40-µm strainer using a plunger and 1-ml syringe filled with sterile 1à PBS. Strained bone marrows cells were washed with PBS and resuspended in 1à sterile PBS with 5% BSA. Afterwards, neutrophils were isolated by using a neutrophil isolation kit (130-097-658, Miltenyi) according to the manufacturerâs instructions. Of cells, 1âÃâ107 were plated onto 150-mm culture dishes in RIPA 1640 (Gibco), supplemented with 10% FBS and 1% penicillinâstreptomycin.
For the generation of NETâDNA, 2âÃâ107 cells (for isolation, see above) were cultured in a 150-mm tissue culture-treated dish. Cells were then stimulated with 100ânM phorbol 12-myristate 13-acetate (PMA) overnight. The next day, supernatant was removed from the culture dish and processed in a centrifugation protocol for isolating NETâDNA. Cell culture supernatants were centrifuged at 500g for 10âmin, then the supernatant was kept and centrifuged again at 15,000g for 10âmin. Supernatant was decanted and the pellet (NETâDNA) was resolved in 50âµl of nuclease-free water. NETâDNA was then labelled with the fluorescent DNA probe DRAQ5 (Alexa 647; Thermo Fisher) following the manufacturerâs instructions.
For ex vivo serum stimulation of neutrophils, 3âÃâ105 cells were cultured in 12-well plates. Cells were then treated with either 25% serum from stroke-operated or sham-operated mice for 4âh. As a positive control, 10ânM PMA was used. Cells were then washed and immediately fixed with 3.7% PFA/sucrose. Fixed neutrophils were then stained with citH3-specific antibody and NETâDNA was counterstained with 1âµM SYTOX green.
cfDNA isolation from human and mouse plasma
Mouse venous blood from cardiac puncture was drawn in 50âmM EDTA 2-ml collection tubes. Samples rested at a maximum of 15âmin at room temperature on the bench. Afterwards, samples were centrifuged at 3,000g for 10âmin at 4â°C. Plasma was isolated, transferred to a new tube and spined again at 3,000g for 10âmin. Plasma was then carefully collected and immediately frozen down at â80â°C until further processing. We used 500âµl plasma to isolate cfDNA with a column-based kit (Plasma/Serum Cell-Free Circulating DNA Purification Kit; 55100; Norgen Biotek). In the last step, DNA was eluted with 30âµl buffer from the column. Afterwards, total circulating DNA and single-stranded DNA were measured with a Nanodrop Spectrophotometer (1000ND, Peqlab). Double-stranded DNA (dsDNA) concentrations were acquired with a Qubit 2.0 fluorophotometer (Invitrogen) using a specific fluorescent dye-binding dsDNA (HS dsDNA Assay kit, Thermo Fisher Scientific). Dilutions and standards were generated following the manufacturerâs instructions.
Length distribution of circulating cfDNA fragments after DNA isolation was acquired using the automated gel electrophoresis platform Bioanalyzer (Agilent) and the High Sensitivity DNA kit (Agilent). Data were analysed using the Bioanalyzer 2100 Expert software (Bioanalyzer, Agilent). Data in Fig. 2h (mouse, 2â72âh after stroke), Fig. 3g (mouse, 6âh after stroke), Extended Data Fig. 7d (mouse, 24âh after stroke), Extended Data Fig. 7e (mouse, 12 or 24âh after myocardial infarction) and Extended Data Fig. 8a (intravenous DNA measured 24âh after injection) were generated following the above protocol.
Human samples from patients with asymptomatic/symptomatic CCA (stroke (human; CCA sample)) were acquired at Hannover Medical School (see âPatient cohort for carotid endarterectomy sample analysisâ). Full-blood samples (blood withdrawal in EDTA collection tubes) were transported to Munich (180âmin or longer transportation time on 4â°C). Once arrived, 3âml of each sample was then centrifuged at 3,000g for 15âmin, and plasma was collected and stored at â80â°C. We used 500âµl plasma to isolate cfDNA with a column-based kit (Plasma/Serum Cell-Free Circulating DNA Purification Kit; 55100, Norgen Biotek). In the last step, DNA was eluted with 30âµl buffer from the column. Details of the cfDNA isolation protocols are in Supplementary Table 4.
Human samples acquired at the medical centre of LMU (Munich, Germany; âstroke (human; cfDNA methylation)â) and at the medical centre of Technical university (Munich, Germany; âMI (human)â) were collected in EDTA containing tubes and centrifuged after 15â30âmin at room temperature. Human myocardial infarction samples were centrifuged at 1,600g for 30âmin, and plasma was collected and stored at â80â°C until cfDNA was isolated. Human stroke samples were centrifuged at 1,500g for 10âmin, and plasma was collected and stored at â80â°C until further processing.
Extracellular vesicle spin down
Mouse plasma samples were diluted 1:1 with nuclease-free water. Dilution was centrifuged at 16,500g for 20âmin, and then supernatant was filtered through 0.22-µm filter. The filtered supernatant was then transferred to an ultracentrifuge and spined at 110,000g for 60âmin. Supernatant was kept as âextravesicularâ DNA. The pellet was resuspended and âvesicularâ DNA concentration was measured.
Human cfDNA methylation pattern analysis
We collected platelet-poor plasma from 17 patients with ischaemic stroke (median of 73.9 years of age, interquartile range of 65.8â87.2 years of age; 10 women (58.8%)) upon hospital admission in the emergency department before any acute treatment or intervention (time from symptom onset to sampling: median of 6.5âh, interquartile range of 2.2â8.3âh). Patients had a median infarct volume of 57.4âml (interquartile range of 40.5â124.0âml).
cfDNA extracted from plasma was treated with bisulfite to convert unmethylated cytosines to uracils, and amplified using a two-step PCR protocol as previously described43, followed by next-generation sequencing. Sequencing results were analysed to determine the proportion of molecules from each locus that carries the methylation pattern of the cell type of interest (typically complete demethylation). We used a cocktail of brain markers (methylation markers of neurons, oligodendrocytes and astrocytes) as previously described44 and another cocktail that amplifies the markers of immune and inflammatory cells: neutrophils, monocytes, eosinophils, and T cells and B cells45. The raw values (GEâmlâ1 and percentage) of DNA methylation analysis are in Supplementary Table 5.
BMDM isolation and cell culture
BMDMs were isolated and cultured as previously described18. In brief, BMDMs were generated from the tibia and femur of transcardially perfused WT mice. After careful isolation and dissection of the tibia and femur, bone marrow was flushed out of the bones through a 40-µm strainer using a plunger and 1-ml syringe filled with sterile 1à PBS. Strained bone marrow cells were washed with PBS and resuspended in DMEMâ+âGlutaMAX-1 (Gibco), supplemented with 10% FBS and 1% gentamycin (Thermo Scientific) and counted. Of cells, 5âÃâ107 were plated onto 150-mm culture dishes. Cells were differentiated into BMDMs over the course of 8â10 days. For the first days after isolation, cells were supplemented with 20% L929 cell-conditioned medium, as a source of M-CSF. Cultures were then maintained at 37â°C with 5% CO2 until they reached 90% or more confluence.
BMDM stimulation with sham or stroke serum
BMDMs were cultured for 8â10 days for full differentiation. Cells were then harvested, washed, counted and seeded in flat-bottom tissue culture-treated 24-well plates at a density of 2âÃâ105 cells per well in a total volume of 500âµl, and then cultured overnight for at least 16âh. BMDMs were then stimulated with LPS (100ângâmlâ1) for 4âh. Afterwards, the cells were incubated with serum from either stroke-operated or sham-operated WT mice (4âh post-surgery) at a concentration of 25% total volume for 1âh. Control-treated BMDMs received only FBS-containing culture medium. Afterwards, cell lysates were collected in RIPA buffer and stored at â80â°C until further processing.
For MMP secretion, the supernatant was discarded after stimulation and the cells were washed with sterile PBS to ensure no leftover serum on the cells. Afterwards, 500âµl serum-free DMEM was added to the BMDMs, which were then incubated overnight for 16âh at 37â°C and 5% CO2. The culture medium was then collected for further analysis.
BMDMs stimulation with NETâDNA
ASCâcitrine (B6.Cg-Gt(ROSA)26Sortm1.1(CAG-Pycard/mCitrine*,-CD2*)Dtg/J) BMDMs were cultured as described above. Cells were then harvested and seeded in a 12-well flat-bottom well plates equipped with 15-mm coverslips. A total of 3âÃâ105 cells were seeded per well in a total volume of 1âml and then cultured overnight for at least 16âh (37â°C and 5% CO2). BMDMs were then primed with LPS (100ângâmlâ1) for 4âh. Afterwards, BMDMs were stimulated with 250âng of DRAQ5-labelled NETâDNA (see the section âNeutrophil isolation and stimulationâ) for 1âh. BMDMs were then washed with PBS and fixed with 3.7% PFA/sucrose. Coverslips containing BMDMs were then removed from the well plates, cytoskeleton was stained for β-tubulin (Thermo Fisher) and nuclei were counterstained with Hoechst 33342 (Immunohistochemistry.com).
En face immunofluorescence staining
Both ipsilateral and contralateral CCAs were carefully dissected, and adventitial fat and ligation nodes were thoroughly trimmed away. CCAs were then cut open, unfolded and pinned out on a silicon-elastomer for fixation in 4% PFA at room temperature for 2âh. The CCAs were then washed for 1âh at room temperature in 5% BSA with 0.3% Triton X-100 (Sigma-Aldrich). Afterwards, CCAs were incubated with rabbit anti-factor XII (1:100; PA5-116703, Invitrogen) at 4â°C overnight. After washing in 5% BSA with 0.3% Triton X-100 for 1âh at room temperature, CCAs were incubated with AF647 goat anti-rabbit (1:100; Invitrogen) and DAPI for 2âh at room temperature. Finally, CCAs were mounted with fluoromount medium (Sigma-Aldrich). Microphotographs were taken with a confocal microscope (LSM 980, Carl Zeiss).
Aspect ratio of collagen I
For assessment of the collagen structural organization at the fibrous cap, 20X PSR images from 3 to 4 sequential segments were taken. Quantitative analysis using ImageJ software was done as previously described46,47. In brief, fast Fourier transformation was performed on the approximately 40-μm subendothelial fibrous cap area. Thresholded fast Fourier transformation images then underwent an elliptic fit and the aspect ratio value was calculated, as a measure of collagen fibre distribution anisotropy at the fibrous cap region.
Statistical analysis and reproducibility
Data were analysed using GraphPad Prism version 6.0. All summary data were expressed as the meanâ±âstandard deviation unless indicated otherwise. All datasets were tested for normality using the ShapiroâWilk normality test. The groups containing normally distributed independent data were analysed using a two-way Studentâs t-test (for two groups) or ANOVA (for more than two groups). Normally distributed dependent data were analysed using a two-way ANOVA. The remaining data were analysed using the MannâWhitney U-test (for two groups) or KruskalâWallis test (H-test; for more than two groups). P values were adjusted for comparison of multiple comparisons using Bonferroni correction or Dunnâs multiple comparison tests. Pâ<â0.05 was considered to be statistically significant.
t-Distributed stochastic neighbour embedding for anti-Ly6G-depleted bone marrow (Extended Data Fig. 8f) was performed using the integrated tSNE platform of FlowJo (Treestar). One thousand CD45+ cells were used per sample. The plugin was set to 250 iterations and a perplexity value of 30.
Principal component analysis (Extended Data Fig. 3f) was performed using Rstudio version 1.1.477. Absolute values from the necrotic core, smooth muscle actin, collagen, fibrous cap thickness and CD68+ macrophage area were Z-scored and then principal components were calculated using the âprcomp()â command (built-in R stats package). The arrows represent the variable correlation showing the relationship between all variables. Principal components were picked by their percentage of explained variance (62.73% (PC1) and 21.05% (PC2)) and visualized using the âggplot2â package (version 3.4.3; https://ggplot2.tidyverse.org). Relative contribution and quality of representation were calculated and visualized using the âcorrplotâ package (version 0.92; https://github.com/taiyun/corrplot).
All in vivo experiments were performed in 3â5 independent experiments. Within one independent experiment, all groups were represented. All in vitro experiments were performed in 2â4 independent experiments. All conditions of the in vitro assays were represented in each independent experiment. Experiment data shown for Fig. 1d,i represent three independent experiments. Experiment data shown in Fig. 2c,e represent three independent experiments. Representative images in Fig. 2f,i,k,m show the outcome of one of three independent experiments. Representative microphotographs in Fig. 3c,e show representative images from experiments that were independently repeated at least five times.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.