Animals
The following mouse lines were used: C57BL/6N (for wild-type control, naive condition and neurosphere assay), Ifnarâ/âIfngrâ/â (for IFNAGRKO naive condition) (B6.Cg-Ifnar1tm1Agt Ifngr1tm1Agt/Atp)63, TiCY (B6-Tg(Nr2e1–creERT2)1Gsc Gt(ROSA)26Sortm1(EYFP)CosFastm1Cgn/Amv, WT-TiCY for wild-type ischaemia condition)43, TiCY-IFN(A/G)R-KO (TiCY-IFN-KO, for IFNAGRKO ischaemia condition) (B6-Tg(Nr2el-CreERT2)lGsc Gt(ROSA)26Sortml(EYFP)CosFastm1Cgn Ifnar1tmlAgt Ifngr1tmlAgt/Amv)63,64, B6 Dnmt3a floxed à VE-Cad CreERT2/4 (Dnmt3a-flox, for AAV_Cre injection experiment)65,66, and TCF-Lef [B6-Tg(TCF/Lef1-HIST1H2BB/EGFP)61Hadj] (WTâTCF-Lef, for cortical and striatal astrocytes)67. Mice were male and were age-matched to 2 months old, except for the âischaemia 3 weeksâ mice (3 months old), the TCF-Lef mice (4 months old) and the Dnmt3a-flox (2â6 months old). No randomization or blinding was performed; see Supplementary Table 2 for a detailed list of experimental conditions and mouse lines. Animals were housed in the animal facilities of the German Cancer Research Center (DKFZ) at a 12âh dark/light cycle with free access to food and water. Humidity was kept at 55% and temperature at 22â°C. All animal experiments were performed in accordance with the institutional guidelines of the DKFZ and were approved by the Regierungspräsidium Karlsruhe, Germany.
Tamoxifen injection and ischaemia
Two-month-old TiCY mice were intraperitoneally injected with tamoxifen. In these mice, tamoxifen-induced Cre recombination takes place in NSCs in the vSVZ, which express TLX (Nr2e1)43, and will stably activate the production of enhanced YFP, labelling NSCs and their progeny. Tamoxifen injection was done as described before18. Two weeks after injection, a bilateral common carotid artery occlusion (BCCAO) injury was performed as described5. Naive mice or injured mice were euthanized at 2 or 21 dpi for single-cell sorting and sequencing, FACS analysis and immunohistochemistry.
Single-cell suspension and FACS
For the ischaemia experiment, the dorsolateral walls of the lateral ventricles (vSVZ) and the striatum were isolated. For the naive experiments, vSVZ, striatum, and olfactory bulb were isolated. Depending on the plate, individual or pooled mice were used to sort single cells on plate. For more information see Supplementary Table 2. Tissues were processed as described previously29 and sorted in a BD FACSAria or FACSFusion at the DKFZ Flow Cytometry Facility. Cells were stained with the following antibodies (all conditions and tissues together): O4-APC and O4-APC-Vio770 (Miltenyi; diluted 1:100), Ter119-APC-Cy7 (Biolegend; 1:100), CD45-APC-Cy7 (BD; 1:200), GLAST (ACSA-1)-PE (Miltenyi:1:50), PSA-NCAM-PE-Vio770 (Miltenyi; 1:75), Prominin 1-A488 (eBioscience; 1:75), and Sytox Blue (Life Technologies, 1:500). For sorting, we size-selected the vSVZ, striatum, or olfactory bulb cells and excluded for doublets, dead cells and CD45+/Ter119+ cells as recently described14. We then sorted different cell populations according to the tissue and experimental condition as follows (also detailed in Supplementary Table 2): For comparison between naive and post-ischaemic conditions: in the vSVZ we sorted GLAST+ cells and O4+ cells, in the striatum we sorted GLAST+ cells. In the olfactory bulb we sorted PSA-NCAM low and high cells. For the ischaemia experiment, we additionally recorded the YFP information for some samples via index sorting. For determining the signal of YFP in the striatum, we specifically performed FACS quantification of naive and ischaemic TiCY mice (2 dpi and 21 dpi). YFP signal was measured from lineage cells in the vSVZ and striatum. Single cells were sorted into individual wells of a 384-well plate. For a small number of plates, we also sorted two different populations of vSVZ astrocytes/NSCs (GLAST+/PROM1+ and GLAST+/PROM1â), PSA-NCAM+ neuroblasts and O4+ oligodendrocytes. For a detailed breakdown of the populations that were sorted, see Supplementary Table 2. When assessing the effects of ischaemia, only GLAST-sorted cells were considered. Flow cytometry data analysis was done with BD FACSDiva 8.0.2,
Miniaturized scNMT-seq protocol
For profiling the transcriptome (cytoplasmic and nuclear mRNAs) and epigenome (DNA methylation and chromatin accessibility) of single cells, we developed and implemented a miniaturized and higher throughput version of the scNMT-seq protocol13. In this new version, the Smart-seq316 method and specific normalization steps were implemented. A detailed version of the protocol is described in17. For some plates (see Supplementary Table 2) we used combinatorial indexing on the genomic DNA fraction, with a multiplexing capacity of 384 cells per run.
Cre virus injection and ischaemia of Dnmt3a
fl/fl mice
To knock out Dnmt3a in cells of the striatum, we stereotactically injected 3âµl of AAV1_P5_Cre virus into both striata (coordinates calculated to bregma: anterior-posterior: 0âmm, medio-lateral: 2âmm, dorsal-ventral: 3âmm). Mice received 5.10âÃâ108 viral genomes in 3âµl. BCCAO was performed in mice at 5 days post-injection and were sacrificed 2 dpi for quantification of PSA-NCAM+ cells. Injected naive mice as well as non-injected naive and post-ischaemic mice were used as control.
For statistical inference, we considered the proportion of PSA-NCAM+ neuroblasts among all live (Sytox Blue negative) cells. Inference was performed using a linear model fitted on the logit scale.
Immunohistochemistry for YFP, DCX and TUNEL
Naive and 2 dpi ischaemic mice were used for immunohistochemistry to quantify YFP+ and DCX+ cells in the striatum (TiCY mice). Briefly, mice were perfused, and brains were fixed overnight with PFA 4%. Immunohistochemistry was performed on 50âµm vibratome sections as previously described29. The following antibodies were used: chicken anti-GFP (AVES, 1:500), guinea pig anti-DCX (Milipore, 1:100), goat anti chicken IgG Alexa 488 (ThermoFisher, 1:500), donkey anti guinea pig IgG Alexa 647 (Jackson Immunoresearch, 1:500). Cell death was analysed by TUNEL staining as described by the manufacturer (B6N mice, Click-iT TUNEL Alexa Fluor Imaging Assay). Nuclei (DAPI) and TUNEL+ nuclei were segmented with Cellpose 2.2.268 and counted in the vSVZ and striatum. The proportion of TUNEL+ cells was averaged across technical replicates (slides) to obtain one value per biological replicate (individual mouse).
Neurosphere assay
To assess the neurogenic potential of striatal and vSVZ cells upon ischaemia, a neurosphere assay was performed on the freshly isolated vSVZ and striatum of naive and post-ischaemic (2 dpi) B6N wild-type mice. Tissue dissociation was performed as previously described29. After dissociation and single-cell preparation, cells were plated in 96-well plates at a density of 1,000 cells in 200âµl per well. After 7 days, the number of wells that had neurospheres were counted. From the set of wells with neurospheres, a maximum of 10 wells were randomly chosen for quantifying the number and size of neurospheres. Three biological replicates were used for every condition.
Processing of single-cell transcriptomic data
Transcriptomic reads were mapped to the mouse genome build GRCm38 (mm10) with STAR 2.7.3a69, using gene annotations downloaded from Ensembl70 Release 102. Both mapping and gene quantification were executed by the zUMIs pipeline 2.9.4f71 as described in the Smart-seq3 protocol (https://doi.org/10.17504/protocols.io.bcq4ivyw).
Processing of single-cell epigenomic data
Genomic reads were first trimmed with Trim Galore 0.4.4 (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) in paired-end mode, and then mapped to GRCm38 with Bismark 0.22.372 in single-end, non-directional mode. After filtering PCR duplicates with Bismark, single-end alignments were merged. DNA methylation at individual cytosines was quantified with Bismarkâs coverage2cytosine script using the NOMe-seq option to distinguish between CpG and GpC contexts.
Quality filtering and analysis of single-cell epigenomic data
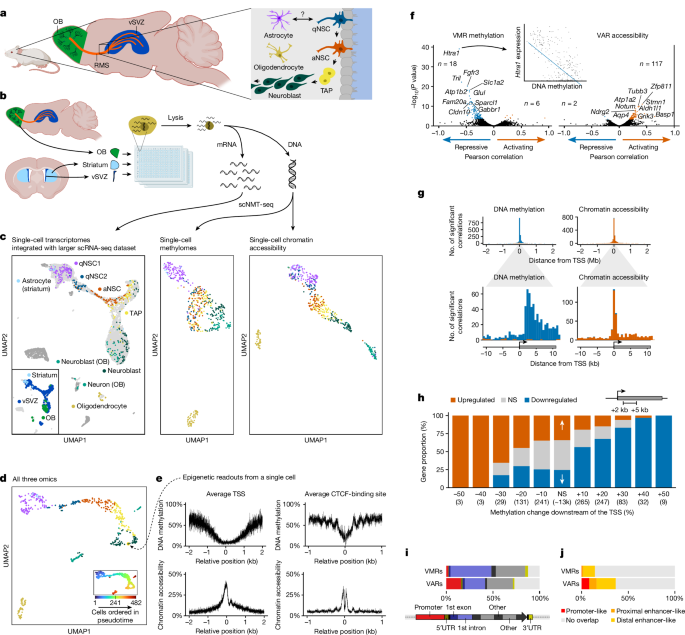
CpG methylation and chromatin accessibility (GpC methylation) was analysed with MethSCAn 0.3.2, a command line tool enabling the analysis of single-cell methylation data that was developed in parallel to this study. For a detailed explanation of the statistical methods, see21. In brief, we used âmethscan prepareâ to separately store CpG and GpC data in an efficient format and to compute quality metrics. Cells with read coverage of less than 50,000 CpG sites, or with poor methylation or accessibility profiles around their TSS were discarded. Methylation and accessibility profiles around TSSs and CTCF-binding sites were computed with âmethscan profileâ. When multiple TSSs were annotated for one gene we selected only the TSS of the âprincipalâ isoform, based on the APPRIS73 score in Ensembl release 102. CTCF-binding sites are based on CTCF ChIP-seq peaks downloaded from the ENCODE portal74 (accession number EENCFF242GNY). We used the âgimme scanâ command of GimmeMotifs 0.15.375 to identify the exact position of CTCF motifs (from JASPAR202276) within these peaks. We furthermore discarded low-quality cells according to a variable threshold on the number of observed genes (minimum 1,500). As reported in prior work using scNMT-seq13,53, some cells passed the RNA quality threshold but did not pass the methylome quality threshold. After filtering, we used âmethscan smoothâ with a bandwidth of 1,000 (500 for GpC data) to quantify the smoothed mean methylation of all high-quality cells over the whole genome. VMRs and VARs were detected with âmethscan scanâ, a sliding window approach that scans the whole genome for regions of high methylation variance between cells. We used a bandwidth of 2,000 (1,000 for GpC data), a step size of 10 and a variance threshold of 0.2. We then quantified methylation and accessibility at VMRs, VARs and promoters (TSSâ±â1,000âbp) using âmethscan matrixâ.
Dimensionality reduction and pseudotime
We used Seurat 4.1.077 to process the scRNA-seq data. To achieve higher resolution, we integrated our transcriptomic data with a much larger scRNA-seq dataset (wild-type cells from18) as previously suggested53. Specifically, after normalizing and finding 3,000 highly variable genes using default Seurat parameters for both datasets, we used FindIntegrationAnchors and IntegrateData using 30 dimensions to integrate the datasets, followed by scaling, principal component analysis (PCA) and UMAP on 30 principal components.
To visualize single-cell methylomes from naive mice, we subjected scaled and centred VMR methylation values to PCA, followed by UMAP on the top 15 principal components, excluding PC 5 which captured cell quality. Since epigenomic data contains missing values, we used a modified PCA that estimates missing values in an iterative manner21. To reduce noise, we used the âshrunken mean of residualsâ reported by âmethscan matrixâ as a measure of methylation, and to reduce technical variation among cells we centred all values for each cell. Only VMRs observed in at least 20% of cells were used for PCA. This threshold did not strongly affect results (Supplementary Fig. 7). Accessibility data was processed in the same manner, with the following differences: Only VARs observed in at least 40% of cells were considered, and promoter accessibility values were used in addition to VAR accessibility.
To project information from all 3 molecular layers into a shared lower-dimensional space, we used MOFA+â1.6.019 aiming for 15 dimensions (factors). As input, we used the expression values of 3,000 highly variable genes (normalized with SCTransform 0.4.178), as well as the same methylation and accessibility values previously used for PCA (promoter methylation and accessibility, VMR methylation, VAR accessibility). We used UMAP on the top 13 MOFA factors, excluding factors 4 and 11, which captured technical variation. We then used Leiden clustering79 on the MOFA factors, followed by slingshot 2.4.080 to obtain pseudotime values informed by both gene expression and epigenetics. Oligodendrocytes were excluded from pseudotime analysis since the focus of our study is on neurogenesis.
The above section describes the analysis of samples from naive mice. To compare naive and post-ischaemic samples, we repeated the dataset integration using all available single-cell transcriptomes from this study and ref. 18 as described above. The same procedure was repeated to integrate additional naive striatal and cortical astrocytes which were sequenced at a later date. Leiden clusters and pseudotime (using slingshot) were re-calculated on the integrated transcriptome PCA.
Correlation of epigenetic features with gene expression
Promoter methylation (mean shrunken residuals) and log-normalized gene expression values were correlated and tested for significance with the R function cor.test (two-sided), using Pearson correlation. VMR methylation was correlated with the expression of the closest gene, as determined with bedtools 2.30.081: bedtools closest -D âbâ -a regions.bed -b gene_bodies.bed. Accessibility was correlated in the same manner. Only regions with genomic reads in at least five cells were considered. Correlation P values were adjusted for multiple testing with the BenjaminiâHochberg method.
We used âmethscan matrixâ to quantify DNA methylation (shrunken mean of residuals) in 3,000-bp-wide intervals downstream of TSSs. Specifically, we considered the TSSs of all protein-coding genes and used the interval from +2âkb downstream to +5âkb downstream of each TSS. TSSs with sequencing coverage in at least 5 cells per group were then tested for differential methylation between astrocytes (vSVZ, striatum) and NSC-lineage cells (qNSC2âneuroblast) with the two-sided Wilcoxon rank sum test. The same approach was applied to log-normalized RNA counts to determine genes up- or downregulated in the NSC lineage. Both sets of Wilcoxon P values were adjusted for multiple testing with the BenjaminiâHochberg method. TSSs were then binned according to the rounded methylation difference, counting TSSs with an adjusted Wilcoxon P valueâ>â0.05 or a methylation difference smaller than 5% as not significant.
ChIPseeker 1.32.082 was used to quantify the number of VMRs and VARs that overlap with gene features, using the options âtssRegion=c(â1000, 1000)â and âoverlap=âallââ. Overlaps with candidate cis-regulatory elements (cCREs, Registry V3, downloaded from https://screen.encodeproject.org/ on 3rd August 202183) were quantified with the mergeByOverlaps function of the GenomicRanges (1.48.0) R package84.
Quantifying methylation change along pseudotime
Correlation heat maps of each molecular layer were generated either by grouping cells by cell state, or by binning cells along pseudotime with a mean of ten cells per bin. For all binned heat maps of non-ischaemic cells, we enforced that each bin only contains cells from one cluster and tissue, so thatâfor example, the first cluster contains only striatal astrocytes. Methylation, accessibility, and expression values were averaged per cell state or bin and the Pearson correlation of all bins was visualized with ComplexHeatmap 2.12.085. We used the ward.D2 method of the R function hclust for hierarchical clustering of cell states. We chose to omit the cell state correlation heat map for chromatin accessibility data since the results depended greatly on the choice of pre-processing methods.
To quantify (de)methylation events in the NSC lineage, we considered all VMRs that were observed in at least 100 cells of the naive wild-type NSC lineage including vSVZ astrocytes. For each VMR, we fit a step function to the methylation values as a function of pseudotime. The function is parametrized by a change point s in pseudotime and two constant values, which the function takes before and after s. Minimizing the sum of squared residuals over this parameter space, we found a most likely value for the methylation change point in pseudotime. VMR change points were considered (de)methylation events if the step function fit was at least 15% better (with respect to the squared residuals sum) than a constant fit without a step. To visualize expression, methylation and chromatin accessibility of genes affected by demethylation in late TAPs (the âsecond waveâ), we selected VMRs with an inferred change point between pseudotime ranks 250 and 400 that intersect with a gene. For each of these VMRs, we visualized VMR methylation, log-normalized expression of its intersecting gene, and VMR accessibility in heat maps.
Epigenetic changes near cell type-specific genes
Representative marker genes for each cell type or stage were determined with the two-sided Wilcoxon rank sum test, by testing log-normalized expression values in cells of interest against the expression values of all other cells. We selected the top 100 most differentially expressed genes among genes with a BenjaminiâHochberg-adjusted P value below 0.05 that also contain a VMR in their gene body. Expression, methylation and accessibility values of these genes and their corresponding promoters or VMRs were averaged.
Transcription factor motif enrichment
Since the PCA on VMR methylation values captured methylation differences between oligodendrocytes and the neurogenic lineage on the first principal component (PC1), we selected the 5,000 VMRs with the highest PC1 loading for oligodendrocyte-specific motif enrichment. We used HOMER 4.461 with the Jaspar2022 motif database76 to identify motifs enriched in these VMRs:
findMotifsGenome.pl VMRs.bed mm10r output/ -len 5,6,7,8,9,10,11,12 -size given -mcheck JASPAR.db -mknown JASPAR.db
The same strategy was used to identify motifs enriched in regions with low methylation in the neurogenic lineage (5,000 VMRs with the highest PC2 loading) and in common parenchymal astrocytes (lowest 5,000 PC2 loadings).
Identification of LMRs and associated GO terms
To identify regions that are differentially methylated between two groups of non-ischaemic cells, we compared VMRs with the two-sided Wilcoxon rank sum test. Only VMRs with genomic coverage in at least 30 cells per group were considered. VMRs with a BenjaminiâHochberg-adjusted P value below 0.05 were labelled LMRs. For visualizing gene expression in volcano plots and heat maps, all LMRs overlapping a gene body were assigned to that gene. We used GREAT 4.0.462 for GO term enrichment of genes near LMRs, using the option âbasal plus extensionâ with a constitutive 20âkb downstream regulatory domain and up to 1,000âkb maximum extension.
To visualize smooth methylation tracks of LMRs and their surroundings, we averaged CpG methylation values in pseudobulk cell groups and smoothed these means with a weighted kernel smoother (tricube kernel, 1,000âbp bandwidth).
Expression and methylation signatures induced by ischaemia
Reactive astrocyte marker gene sets46,47 were used to calculate expression signatures (mean log-normalized expression of the respective gene set). We used the âPan reactiveâ, âA1 specificâ and âA2 specificâ gene sets from ref. 46. We used the lipopolysaccharide-induced, cluster-specific differential expression results reported in ref. 47 to determine differentially expressed genes that are either shared among all astrocyte clusters (consistently lipopolysaccharide-induced), or only in one specific cluster (as in fig. 3c in ref. 47). NSC methylomes and astrocyte methylomes were distinguished based on the mean methylation of all astrocyte and NSC LMRs; the depicted methylation score is the difference of these two means.
To detect DMRs induced by ischaemia, we selected all qNSC2 cells, aNSCs and TAPs from the vSVZ. We then used the command âmethscan diff –bandwidth 2000 –stepsize 100 –threshold 0.05 –min-cells 6â to test cells from 2 dpi against cells from naive mice21. This approach was repeated to test 21 dpi cells against naive. Data exploration and visualization was done in R/tidyverse 1.3.1.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.