Animals
Opossums and mice (C57BL/6J and Mus spretus) were maintained in the Francis Crick Institute Biological Research Facility in accordance with UK Animal Scientific Procedures Act 1986 regulations (project licence P8ECF28D9) and subject to Francis Crick Institute’s internal ethical review. Additional opossums were housed at the University of Texas Rio Grande Valley under Institutional Animal Care and Use Committee protocol AUP-19-31. No randomization or blinding was performed and no statistical methods were used to predetermine sample size. Mice were housed in individually ventilated cages (GM500, Tecniplast) with a 12:12-h light/dark cycle, a temperature of 20–24 °C and humidity of 55% ± 10%. Adults (8 weeks–6 months) were housed in groups of 3–4 animals, with the different sexes housed separately. Mice had free access to water and food and were provided enrichment activities including rodent balls and nesting boxes. Matings for timed collection of embryos were conducted by placing female mice into the cage of male mice at approximately 17:00. Observation of a vaginal plug the following morning was taken to indicate that mating had occurred.
Opossums were individually housed in Double Decker cages (GR1800, Tecniplast), with male and female animals housed in separate rooms except during mating periods. The temperature of the housing was maintained between 24 °C and 28 °C, and humidity was maintained between 55% and 75%, with a 14 h/10 h light/dark cycle. Opossums had free access to dried food and water, supplemented every second day with live mealworms, and weekly by fresh fruit. To induce oestrous before mating, adult male and female (≥6 months) mice were placed in single-storey rat cages immediately adjacent to each other for 2 days, and then swapped into each other’s cages for an additional 2 days. Subsequently, pairs were placed into the same cage and kept together for 10 days, during which period animals were monitored by infrared CCTV camera for mating behaviour69.
Collection of gamete, embryo and tissue samples
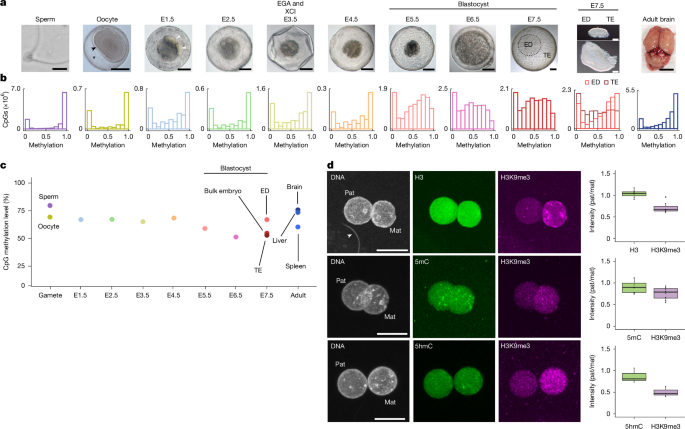
Mouse E0.5 (11 h post coitum (hpc)) and E3.5 (82–84 hpc) embryos were recovered by flushing the uteri with PBS (Gibco number 14190-094) from a blunt-ended needle under a Leica MC80 dissecting microscope, aspirated using a Stripper pipette with a 275-μm tip (MXL3-STR and MXL3-275, Cooper Surgical), and washed three times through drops of clean PBS. For immunofluorescence, zygotes were fixed in 4% PFA in PBS with 0.2% Triton X-100 for 60 min at room temperature and washed three times in 0.2% Triton X-100 in PBS with 0.01% polyvinyl alcohol (PVA; number P8136, Sigma). For sequencing of blastocysts, the zona pellucida was removed by incubation in acid Tyrode’s solution (T1788, Sigma-Aldrich), and then the embryo was washed three times in PBS, snap-frozen on dry ice and stored at −80 °C until processing. Picking-buffer-only negative controls were collected in parallel, and processed through the library preparation procedure to verify the absence of contamination.
Opossum embryos were recovered from the uteri in PBS under a Leica MC80 dissecting microscope at E0.5 (29 hpc), E1.5 (36 hpc), E2.5 (60 hpc), E3.5 (84 hpc), E4.5 (108 hpc), E5.5 (132 hpc), E6.5 (156 hpc) and E7.5 (180 hpc). In the case of oocyte collection, the female animal was mated to a vasectomized male animal, and oocytes were recovered from the uterus at 36 hpc. Embryos were aspirated using a Stripper pipette with a 600-µm tip (MXL3-600, Cooper Surgical), or a micropipette fitted with a 200-µl tip for large E7.5 embryos, and washed three times in PBS. Samples were imaged using a Leica DMIL LED microscope with a 20× objective and then further processed for sequencing or immunofluorescence. The shell coat was perforated with 1-μm dissecting needles (10130-20, FST) and incubated in 5 mg ml−1 protease in PBS (P8811-100MG, Sigma) at 32 °C for between 2 and 7 min. The sample was transferred into fresh PBS, and any remaining mucoid coat was removed by disaggregation with dissecting needles.
For sequencing, the oocytes and E0.5–7.5 embryos were washed through three drops of fresh PBS, dispensed to 0.2 µl tubes, and snap-frozen on dry ice and stored at −80 °C until further processing. We collected multiple single embryos for each time point, and in addition collected pooled litters of oocyte, E2.5 and E3.5 samples (Supplementary Table 1). For E7.5 embryonic disc and trophectoderm samples, after removal of the shell coat, the embryonic disc was isolated from the trophectodermT by dissection with 1-μm needles, washed through three drops of PBS, and snap-frozen as above. For single-cell sequencing, embryos were washed through three drops of fresh PBS with BSA, and incubated in TrypLE (12604013, ThermoFisher) diluted to 0.5× in PBS for 2–8 min at 35 °C. Following TrypLE incubation, embryos were moved to PBS with BSA and disaggregated to single cells by repeated pipetting through a narrow-bore pipette, before nucleosome, methylome and transcriptome (NMT)-sequencing processing. Picking-buffer controls were included as described above. For immunofluorescence, E0.5 zygotes were fixed and washed as above for mouse embryos.
For collection of sperm from mouse and opossum adult male animals, epididymides were dissected from the testes and rinsed in PBS. Cauda epididymides were transferred to 2 ml Bigger–Whitten–Whittingham buffer (opossums) or TYH buffer (mice). Several small incisions were made, followed by incubation at 37 °C for 30 min to facilitate sperm swim out. The swim-out was diluted approximately 1:10 in PBS, and visualized on a dissecting microscope using a dark field. Individual sperm were aspirated using a Stripper pipette with a 100-µl tip (MXL3-100, Cooper Surgical) and washed three times in PBS before collection in 5 µl RLT Plus (1053393, Qiagen) containing 2% β-mercaptoethanol. For collection of pools of sperm, multiple sperm cells were picked in one pipette and processed together through washing, collection and freezing as for single sperm cells. Picking-buffer controls were included as described above.
For collection of genomic DNA from adult mice, and genomic DNA and RNA samples from adult opossums, brain, liver and spleen tissues were dissected into ≈10-mm pieces, snap-frozen in liquid nitrogen, and stored at −80 °C. To facilitate allele-specific analyses, a cross was set up with parent animals from the LL2 opossum stock, and tissue samples were collected from the parents for whole-genome sequencing (WGS) and genomic variant identification, and from three male and three female littermates for BS-seq and RNA-seq. Mouse tissue samples were collected from a model exhibiting skewed XCI through a targeted Xist deletion70 (that is, C57BL/6J Xisttm1Jae × Mus spretus F1 hybrid cross). We collected three heterozygous female offspring in which XCI is completely skewed, and three male littermates with Xist deleted. Frozen tissue was pulverized using a pestle and mortar pre-cooled on dry ice, and used for genomic DNA extraction with the PureLink Genomic DNA Mini kit (K18290-02, Invitrogen) or RNA extraction using the Ambion RNAqueous-Micro Kit (AM1931, ThermoFisher). Samples were then processed for WGS, BS-seq or RNA-seq library preparation.
BS-seq
Library preparation was performed with oligonucleotides compatible with the NEBNext library preparation kit (E7535S) following an established method71 and described briefly here. For gamete and embryo samples, samples were lysed in 2.5 µl RLT Plus and processed following the low-input library method71. For brain, liver, spleen and fibroblast samples, 6 ng of genomic DNA was used to prepare libraries following the bulk method71. Samples were spiked with 6 fg of Lambda DNA (D152A, Promega) and bisulfite-converted using the EZ Methylation Kit (D5020, Zymo). Bisulfite-converted DNA was purified using the PureLink PCR Purification Column Kit (K310050, Invitrogen) with an additional treatment with M-desulfonation buffer (EZ Methylation Kit, Zymo). Samples were eluted into a mixture containing 0.4 µM Preamp primer (5′-CTACACGACGCTCTTCCGATCTNNNNNN-3′) and submitted to one (brain, liver and spleen samples) or five (gamete and embryo samples) rounds of pre-amplification with Klenow exo− (M0212M, NEB). Unused oligonucleotides were degraded by incubation with exonuclease I (M0293L, NEB), and samples were purified with Ampure XP beads (A63881, Beckman Coulter). Samples were resuspended in a mixture containing 0.4 µM Adaptor 2 Oligo for NEB indices (5′-CAGACGTGTGCTCTTCCGATCTNNNNNN-3′) and adaptor-tagged by incubation with Klenow exo−. Samples were purified using Ampure XP beads and resuspended in a mixture containing 0.2 μM NEBNext Universal Adaptor and 0.2 μM NEBNext Index Adaptor (E7535S, NEB), and library amplification was performed using KAPI Hifi HotStart polymerase (KK2502, KAPA Biosystems). Varying numbers of PCR cycles were performed depending on input amount (opossum oocytes and E1.5–E5.5 embryos: 19 cycles; mouse E3.5 embryos and opossum E6.5 and E7.5 embryos: 10–14 cycles; sperm: 10–18 cycles; brain, liver and spleen: 10 cycles). Library sequencing was carried out by the Francis Crick Institute Advanced Sequencing Facility (ASF). Gamete and embryo libraries were sequenced (100-base-pair (bp) paired end) on an Illumina HiSeq 4000, yielding between 47 million and 337 million reads per library. Brain, liver and spleen libraries were sequenced (150-bp paired end) on an Illumina HiSeq 4000, yielding between 198 million and 363 million reads per library.
RNA-seq
Purified RNA was submitted to the Francis Crick Institute ASF for preparation of cDNA using the SMART-Seq v4 Ultra Low Input RNA Kit (634894, Takara), followed by library preparation using the Nextera XT DNA Library Preparation Kit (FC-131-1096, Illumina). Sequencing (100-bp paired end) was performed on an Illumina HiSeq 4000, generating between 54 million and 156 million reads per library.
NMT-seq
Disaggregated single cells were deposited into individual wells of a 96-well plate, and NMT-seq libraries were prepared as previously described71. In brief, cells were incubated with M.CviPI for 15 min, followed by separation of mRNA and genomic DNA using Oligo-dT beads, and subsequent preparation of RNA-seq and BS-seq libraries. Sequencing (100-bp paired end) was performed on an Illumina HiSeq 4000.
DNMT1-knockout opossum fibroblasts
Primary opossum fibroblasts were derived from a newborn male animal and immortalized using SV40-tag virus infection. Single-cell clonal selection was performed to identify an euploid cell line. Opossum fibroblasts were maintained in DMEM (Gibco) supplemented with 20% fetal bovine serum, 1% GlutaMax (Gibco), 1% sodium pyruvate (Gibco) and 1% penicillin–streptomycin (Gibco, 10,000 U ml−1) and were routinely tested and found to be negative for mycoplasma. Single-guide RNAs (sgRNAs) targeting all opossum DNMT1 paralogues (DNMT1A, DNMT1B and DNMT1Ψ) were designed using the online tool CRISPRdirect (https://crispr.dbcls.jp/): DNMT1 gRNA 1: 5′-TCTGAAGGCTTTCATCAAGC-3′; DNMT1 gRNA 2: 5′-CATTGTGGGCCATTGAAATG-3′. sgRNAs were annealed and ligated into the targeting plasmid px333-puro. The plasmid px333-puro was obtained after cloning a puromycin-resistance cassette isolated from px459v2 (gift from F. Zhang, Addgene number 62988) into the px333 vector (gift from A. Ventura, Addgene number 64073)72.
Immortalized opossum fibroblasts were seeded onto gelatin-coated wells of 6-well plates. The following day, fibroblasts were transfected using PEI MAX (49553-93-7). A 2 µg quantity of plasmid (with sgRNAs or empty plasmid for negative control) was added to 200 µl of Opti-MEM (Gibco) and 8 µl of PEI MAX (1 mg ml−1). One day after transfection, puromycin (2.5 µg ml−1) was added for 48 h to select successfully transfected cells. Control and DNMT1-knockout cells were fixed for RNA fluorescence in situ hybridization (RNA FISH) or frozen down for quantitative PCR with reverse transcription (qRT–PCR) at specific time points.
RNA FISH
Cells were washed in cold PBS and treated with ice-cold permeabilizing solution (0.5% Triton X-100, 2 mM vanadyl ribonucleoside complex in PBS) for 10 min. After fixation using ice-cold 4% PFA in PBS for 10 min, cells were rinsed in ice-cold PBS twice, dehydrated through ice-cold 70%, 80%, 95% and 100% ethanol for 3 min each, and air-dried. BAC VM-18-303M7 (CHORI) was used for RSX RNA FISH. BAC DNA was labelled using Nick Translation Kit (Abbott) with fluorescent nucleotides (spectrum orange-dUTP; 02N33-050, Abott), and cells were hybridized with a denatured mix of probes along with 1 µg salmon sperm DNA in hybridization buffer (50% formamide, 10% dextran sulfate, 1 mg ml−1 PVP, 0.05% Triton X-100, 0.5 mg ml−1 BSA, 1 mM vanadyl ribonucleoside complex in 2× SSC) at 37 °C overnight in a humid chamber. Stringency washes were performed on a hot plate, three times for 5 min in 50% formamide in 1× SSC (pH 7.2–7.4) preheated to 45 °C, and three times for 5 min in 2× SSC (pH 7–7.2) preheated to 45 °C. Cells were mounted in antifade containing DAPI (Vector) with a coverslip and stored at −20 °C.
RNA purification and qRT–PCR
RNA from control and DNMT1-knockout male opossum fibroblasts across three different time points and three replicates was purified using RNAqueous-Micro Total RNA Isolation kit (Invitrogen, AM1931). Purified RNA was retrotranscribed using the Maxima First Strand cDNA synthesis kit (Thermo Scientific, K1641). The following primers were used to assess gene expression in RT–PCR assays using PowerUp SYBR Green (Applied Biosystems, A25780): RSX (5′-AGAAGGGACCCCAAGACAC-3′, 5′-TGGGTCACTTCCACTTCCTC-3′); DNMT1 (5′-GACGCAGTAACACTGGAGCA-3′, 5′-ATCCCATTCCAACCTTCCAT-3′); H19 (5′-TCCAGCAGCAGTCAGTGAAC-3′, 5′-TCATCCATCCATGAGCAGAG-3′); ABCD4 (5′-ATCGATAATCCGGACCAGCG-3′, 5′-ATGATCAGCTTGCTGGCCAT-3′), GAPDH (5′-TAAATGGGGAGATGCTGGAG-3′, 5′-ATGCCGAAGTTGTCGTGAA-3′).
Fibroblast bulk RNA-seq
Libraries from control and DNMT1-knockout RNA samples from day 4 and 8 were prepared with the NEBNext Ultra II Directional PolyA mRNA kit according to the manufacturer’s instructions. Libraries were sequenced on the Illumina NovaSeq 6000 system (paired end, 100-bp read length). Raw RNA-seq reads were processed using the RNA-seq nf-core pipeline (v3.2); star_rsem was used to generate raw reads counts. The read counts were processed in R using the DESeq2 package (v1.36). Genes expressed at very low levels were filtered out by applying a rowSums filter of ≥5 to the raw counts table. Raw counts were normalized using the DESeq() function, specifying ~genotype_day in the design formula. log2[fold change] and adjusted P values between DNMT1 knockout and control were calculated using the lfcShrink() function in DESeq2, specifying type = ‘ashr’, analysing day 4 and day 8 separately.
X/A ratios were calculated using the median expression of X and autosomal genes in each sample after filtering out genes expressed at low levels (transcripts per million (TPM) of <1). X/A ratios between control and DNMT1-knockout samples were compared by Student’s t-test. Repetitive element expression was analysed as above for opossum embryos.
WGS
Samples of 3 μg of genomic DNA prepared from ear snips from one male and one female opossum from the LL2 stock were submitted to the Francis Crick Institute ASF for library construction (KAPA HyperPlus). Libraries were sequenced (150-bp paired end) on a HiSeq 4000, producing 256,834,288 reads (99.23% mapped) for the female animal and 122,371,579 reads (99.27% mapped) for the male animal. Data were used for identification of genomic variants, described below.
Preparation of a modified MonDom5 reference genome and annotations
We modified the MonDom5 reference genome to include a gap-filling long-read sequence of the RSX locus73. We prepared modified gene, repeat and CGI annotation files with corrected coordinates on the gap-filled X chromosome. We also included annotation for both opossum DNMT1 paralogues17 in our modified gene annotation file.
Expression analysis of opossum DNA methylation factors and repetitive elements
Opossum embryo RNA-seq data23 and DNMT1-knockout RNA-seq data were mapped to the modified MonDom5 reference genome and ASM229v1 reference genome, respectively, using HISAT2 (ref. 74) with the command hisat2 -3 0 -5 9 –fr –no-mixed –no-discordant, and read summarization at genes was performed using the Rsubread package75, excluding multi-mapping reads. Fragments per kilobase of transcript per million mapped read (FPKM) values were calculated using the scater package76. Read summarization at repetitive elements and calculation of counts per million were performed using Telescope77. Line plots showing mean and standard error of gene and repeat expression were generated using ggplot2.
Analysis of methylation at repetitive elements
To generate a bigwig file per time point, the filtered methylKit csv files were converted from csv to bedGraph using awk and then converted to bigwig using the UCSC bedGraphToBigWig utility. Genomic coordinates of the transposable elements, specifically L1, MIR and ERV1, were obtained from the mondom5 RepeatMasker GTF file, selecting for those in the ‘forward’ orientation. Using deepTools78, the computeMatrix function in scale-regions mode was used to calculate the methylation score, using the parameters –binSize 50 –averageTypeBins mean -a 1000 -b 1000. The plotProfile function was used to generate the profile plot for each transposable element, using the parameters –perGroup –yMin 0 –yMax 1.
Methylation segmentation of E7.5 embryonic disc and trophectoderm
The methSeg function from the R package MethylKit79 was used to divide the genome into contiguous stretches of similar methylation level in E7.5 embryonic disc and trophectoderm samples. Parameters used were minSeg = 5, G = 1:3, join.neighbours = TRUE. Individual CpG sites with a minimum coverage of five reads in both samples were used for the analysis. The length and average methylation level of each segment were plotted using ggplot2.
Single-cell methylation analysis
Single-cell RNA-seq data were aligned to the opossum ASM229v1 genome, using the nf-core rnaseq pipeline (3.2), using the parameters –aligner star_rsem –bam-csi-index. Raw counts were loaded into Seurat (4.3.0)80 for analysis in R (4.2.2). In total, there were 74 E5.5 samples, 142 E6.5 samples and 42 E7.5 samples. Each dataset was normalized using NormalizeData, and then datasets were integrated together using the functions SelectIntegrationFeatures, FindIntegrationAnchors and then IntegrateData. The integrated dataset was scaled used ScaleData. Principal component analysis was applied using RunPCA. Uniform manifold approximation and projection was estimated for the integrated dataset using RunUMAP. Module scores for EPI and trophectoderm were calculated using the AddModuleScore function, with the following genes used as markers for each cell lineage. EPI: NANOG, PRDM14, POU5F1 and POU5F3; trophectoderm: GATA2, GATA3, TEAD4, AQP3 and KLF4.
Single-cell BS-seq data were trimmed using the TrimGalore!81 command trim_galore –clip_R1 6 –three_prime_clip_r1 6, mapped using Bismark82 with the command bismark –non_directional –un –ambiguous –multicore 2 and deduplicated using the command deduplicate_bismark. Methylation information was extracted with the command bismark_methylation_extractor –comprehensive –multicore 2 –bedGraph –CX –cytosine_report –nome-seq. Average CpG methylation was calculated per cell and plotted according to the cell lineage determined for the corresponding RNA-seq library (module score, above).
Analysis of methylation in eutherian oocytes
Raw RNA-seq and BS-seq data were processed using the nf-core rnaseq (3.12) and methylseq (2.5.0) pipelines, respectively. Gene bodies and intergenic regions for each organism were identified using the GenomicFeatures83 R package. Following a methodology similar to that in ref. 34, genes were classified as active if their TPM was >5 and inactive if their TPM was ≤1, and Bismark CpG methylation calls were imported into R using the methylKit79 R package, then destranded, pooled by sample condition and filtered for CpG sites with a minimum coverage of three reads. The regionCounts function was used to calculate the methylation level across active genes, inactive genes and intergenic regions. These were visualized as violin plots used ggplot2 (ref. 84).
Identification of genomic variants
WGS data from LL2 parent opossums were used to identify genomic variants. WGS reads were trimmed using TrimGalore!81 with the command trim_galore –cores 4 — paired –fastqc –gzip –retain_unpaired –clip_R1 10 –clip_R2 10 — three_prime_clip_R1 5 –three_prime_clip_R2 5. Libraries were mapped to the MonDom5 reference genome using BWA-MEM85 with the command bwa mem -t 32 -M -R. Paired and unpaired mapped reads were then merged, sorted and indexed using SAMTools86. Variants were called using the GATK best practices pipeline87. For the base recalibration step, known variants were not available for opossums. Therefore, variants were initially called independently with three pipelines: BCFtools88, Varscan89 and GATK87. Variants identified by all three pipelines were considered high-confidence variants and were used for GATK base recalibration. Subsequently, variants were called for each opossum, and then combined, and genotypes were annotated to produce a variant call file. BEDtools maskfasta was used to create an N-masked version of the MonDom5 reference genome from the complete set of 25 million variants. Using a custom R script, variants were filtered to include only hemizygous and heterozygous single nucleotide polymorphisms (SNPs). The resultant 2 million SNPs were used in the SNPsplit pipeline90.
For mice, genomic variant data (41,668,158 SNPs) were derived from the C57BL/6J and M. spretus genomes (Mouse Genomes Project91), and the program SNPsplit90 was used to generate an mm10 reference genome in which parental genomic variants were N-masked.
Methylation analysis
BS-seq reads were trimmed using the TrimGalore!81 command trim_galore –clip_R1 6 –three_prime_clip_r1 6. Reads were mapped to the mm10 (mouse) or MonDom5 (opossum) reference genome using Bismark82 on single-end mode with the command bismark –non_directional –un – ambiguous. For adult libraries, reads were mapped to the N-masked genomes, and bam files were generated for all mapped reads as well as for allele-specific reads using SNPsplit90 with the command SNPsplit –bisulfite –conflicting. Library statistics (Supplementary Table 1) were extracted from TrimGalore! and Bismark output reports. Bam files were deduplicated and methylation calls were extracted using Bismark. CpG methylation calls were imported into R92 using the package methylKit79, and principal component analysis was performed using all CpG methylation calls for each individual sample. Data were destranded and pooled by sample condition, and the number of CpG sites captured at different coverage thresholds was calculated. For subsequent analyses, data were filtered for CpG sites with a minimum coverage of five reads. Methylation distribution histograms, mean methylation plot and genomic coverage plots were generated using ggplot2 (ref. 84). This workflow was independently reproduced to ensure that the results were accurate and robust.
Analysis of gamete DMRs
DMRs between gametes were identified as follows: 100-bp non-overlapping tiles covered by a minimum of three reads in both oocyte and sperm samples and containing a minimum of one CpG in a tile were identified. The methylation level of each tile was compared between oocyte and sperm using the diffMeth function from the R package methylKit79, with the parameters difference = 80 and qvalue = 0.01. Gamete DMRs with putative transient or life-long retention of differential methylation were defined as tiles with intermediate methylation levels (40−60% methylated), in either E3.5 and E7.5 embryonic disc or trophectoderm (transient embryonic or trophectoderm DMRs), or in E3.5, E7.5 embryonic disc, and all three of brain, liver and spleen (life-long DMRs). The gene nearest to each life-long DMR was identified using the nearest function from the R package GenomicRanges83, and this list was manually checked for previously reported marsupial imprinted genes29,93. To retain maximum read coverage in low-input embryo samples, all libraries per time point were in silico-pooled for this analysis. The dataset therefore included male and female embryos, precluding analysis of sex differences or the X chromosome.
Sex-specific methylation analysis
CpG methylation calls were destranded, pooled by sample condition (sex and tissue) and filtered for minimum coverage of five reads in all conditions. For embryo samples, sex was inferred from the ratio of reads mapping to chromosome X and pseudo-Y (coding sequence of 19 known opossum Y-chromosome genes)94,95. Autosomes and X-chromosome allelic methylation distributions were represented as ridgeline plots using the R package ggridges96.
Allele-specific methylation analysis
Allele-specific CpG methylation calls were destranded, pooled by sample condition (sex, tissue and parental genome) and filtered for minimum coverage of five reads in all conditions. To avoid loss of X-linked sites in female samples due to low coverage of the X chromosome in male files, data import and filtering for X chromosomes was performed separately with the coverage parameters as for autosomes, but excluding paternal genome files from male samples. Ridgeline plots were generated using ggridges96 as above.
Escape-gene methylation analysis
To generate a list of genes expressed in each tissue, RNA-seq reads were trimmed using TrimGalore!81 with the command trim_galore –paired — clip_R1 10 –clip_R2 10. Trimmed reads were aligned to the N-masked MonDom5 reference genome using HISAT2 (ref. 8) with the command hisat2 –no-softclip –no-mixed –no-discordant. Mapped files were converted to bam files and merged by sample using SAMtools86. Reads overlapping annotated genes were quantified using the command featureCounts from the R package Rsubread75, excluding multi-mapping reads. Files were merged by biological replicate, and FPKM values were calculated using the R package DESeq2 (ref. 97) using the robust median ratio method. Gene models annotated as pseudogenes were excluded, and a threshold of FPKM > 1 was imposed. Expressed genes were categorized as escaping or subject to XCI on the basis of published work45. Methylation level at genes was calculated using the methylKit functions regionCounts and percMethylation, and represented as violin plots for each category using ggplot2 (ref. 84).
Immunofluorescent staining of mouse and opossum embryos
Embryos were permeabilized in 0.5% Triton X-100 in PBS at 4 °C overnight, followed by three washes in 0.2% Triton X-100 in PBS with 0.1% PVA (0.2% TX–PBS–PVA). After 3.5 M HCl treatment for 30 min at room temperature, embryos were washed three times in 0.2% TX–PBS–PVA, blocked for 1–4 h at room temperature in 3% BSA in 0.2% TX–PBS–PVA (0.22-µM-filtered, blocking solution) and incubated overnight at 4 °C in primary antibody in blocking solution. Antibodies used were 5mC (BI-MECY-0100, Eurogentec) at 1:100, 5hmC (75-268, NeuroMab) at 1:1,000, H3K9me3 (07-442, MerckMillipore) at 1:200, H3 (ab1791, Abcam) at 1:100. Embryos were washed three times in 0.2% TX–PBS–PVA and incubated in Alexa Fluor-conjugated secondary antibodies at 1:250 in blocking solution for 2 h at room temperature. Three further 0.2% TX–PBS–PVA washes were performed, and DNA was counterstained with 10 µg ml−1 propidium iodide (P4170, Sigma) for 1–2 h at room temperature. Samples were washed three times with PBS with 0.1% PVA and mounted in Vectashield (H-1000-10, Vector Laboratories). Samples were imaged using an LSM710 confocal microscope with 1-µm z-sections. Images were processed and fluorescence intensity was measured using Fiji98.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.