Mice
For LD50 and inhibitor experiments, C57BL/6 female mice from Jackson Laboratories (000664) were used. Young mice were used at 12 weeks of age or roughly 12 weeks of age (12–14 weeks). Aged mice were used at 72–76 weeks of age. Trim63 mutant mice were generated by Cyagen using CRISPR–Cas mediated genome engineering to delete exons 1–8. The mutant line was maintained as heterozygous and Trim63+/− and Trim63−/− were used as Trim63 knockout in our study. The Trim63+/+ generated from the heterozygous breeding scheme were used as wild-type controls. Foxo1 mckcre mice were a generous gift from M. Febbraio (Monash University) and were bred in our colony with the following breeding scheme: Foxo1 mckcre+ × Foxo1 mckcre−. cre+ and cre− littermates were used for experiments. Foxo1 Myh6cre mice were generated from a series of crosses using our floxed Foxo1 mice to B6/FVB-Tg(MyH6-cre_2182Mds/J) from Jackson Laboratories (011038). All experiments were performed in our AAALAC-certified vivarium, with approval from The Salk Institute Animal Care and Use Committee (IACUC). In accordance with our IACUC guidelines, in an effort to reduce the number of animals used for our experiments, where possible and appropriate, experiments were done in parallel and shared controls were used. This is indicated in the figure legends where relevant. Mice were housed with a 12-h light/dark cycle, humidity 30–70% and temperature of 18–26 °C (64–79 °F).
Bacteria
Bacteria used were E. coli O21:H+ (ref. 43) and S. aureus (American Type Culture Collection strain 12600). For a list of complete key resources used in this study, refer to Supplementary Table 1 in the Supplementary Information.
Method details
Culturing E. coli O21:H+ and S. aureus for mouse infection
E. coli O21:H+ was incubated overnight at 37 °C on an eosin methylene blue plate treated with ampicillin sodium salt (1 mg ml−1), vancomycin hydrochloride (0.5 mg ml−1), neomycin sulfate (1 mg ml−1) and metronidazole (1 mg ml−1) antibiotics to grow single colonies. S. aureus was incubated overnight on a Luria-Bertani (LB) plate at 37 °C without antibiotics for colony growth. The next day, a single colony of E. coli O21:H+ was inoculated into LB-AVNM media. A single colony of S. aureus was inoculated into LB without antibiotics. Both cultures were shaken overnight at 37 °C (250 rpm). The following morning, the optical density was measured and an inoculum with a 1:1 mixture of the bacteria was prepped with the appropriate amount of both bacteria in sterile 1× PBS that was used directly for mouse infections.
Mouse infection models
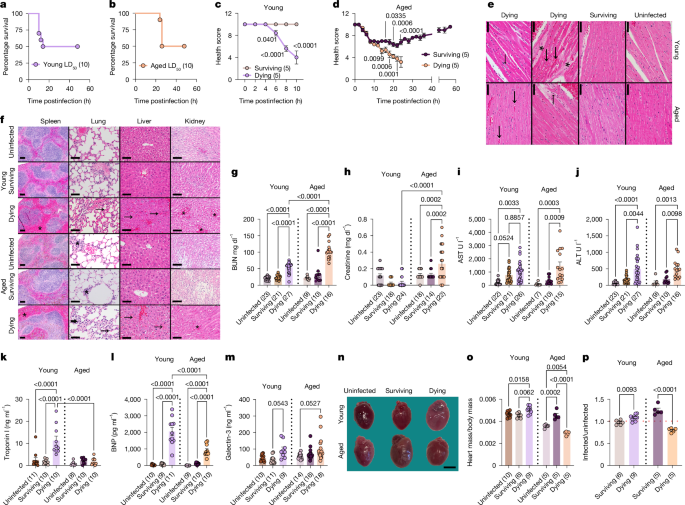
Mice were infected intraperitoneally with the appropriate dose of bacteria and put back into their home cage. Mice were group housed for the experiments. For LD50 experiments the dose of total bacteria used was 1 × 108 CFU. This dose was titrated up or down for low dose and high dose models. Inoculums were serially diluted and plated to confirm the infectious doses. Immediately after infection, food was removed for the first 10–12 h postinfection to control for any potential variations in the sickness-induced anorexic response. Mice were clinically monitored as described below every 2 h postinfection. For some experiments, mice were clinically monitored every 2 h for the first 10–12 h postinfection and then again at 24 h. For experiments involving inhibitors, the details are provided below. Mice that reached clinical endpoints were euthanized according to our animal protocol.
Survival
Mice were clinically monitored as described below every 2 h postinfection. For some experiments, mice were clinically monitored every 2 h for the first 10–12 h postinfection and then again at 24 h. Mice that had to be euthanized because they reached clinical endpoints during the infection, in addition to those that succumb to the infection, were included in our survival counts.
Rectal temperature
Rectal temperatures were taken every 2 h postinfection for the first 10–12 h, and then every 24 h as noted using the Digisense Type J/K/T thermocouple meter. Temperatures are shown as temperatures over time, temperatures at a defined time point or the minimal temperature shown by the animals over the course of the infection. This is indicated in the figure legends. Rectal temperatures were also used to calculate health scores to generate health trajectories.
Grading system for monitoring morbidity
We use the following morbidity scale to quantify the morbidity of mice. Infected mice are clinically assessed using this morbidity scale every 2 h postinfection. For some experiments, mice were clinically monitored every 2 h for the first 10–12 h postinfection and then again at 24 h. Morbidity scores are shown as scores over time, scores at a defined time point or the maximal morbidity shown during the infection (the lower the score, the greater the morbidity). This is indicated in the figure legends. Morbidity scores were also used to calculate health scores to generate health trajectories.
(5) Normal. Normal exploratory behaviour, rearing on bind limbs and grooming.
(4) Mild. Reduced exploratory behaviour, rearing on bind limbs and grooming. Slower and/or less stead gait, but free ambulation throughout the cage.
(3) Moderate. Limited voluntary movement. Slow, unsteady gait for less than 5 s.
(2) Severe. No voluntary movement, but mouse can generate slow, unsteady gait for more than 5 s.
(1) Moribund. Mouse does not move away from stimulation by research but can still right itself.
(0) Deceased.
Generating health trajectories
Rectal temperatures were assigned bin scores based on the following strategy. The sum of the temperature bin score (below) and morbidity score (above) for each mouse at each time point was determined to generate health scores. Mice that were deceased or were euthanized because they reached clinical endpoint were assigned a score of 0. Health scores were then plotted against time to generate the health trajectories. Temperature bin scores: (5) 35–>38 °C, (4) 31–34.9 °C; (3) 28–30.9 °C, (2) 25–27.9 °C, (1) 22–24.9 °C and (0) lower than 22 °C.
MRI
An Echo magnetic resonance imaging (MRI) machine and Echo MRI body composition software v.2008.01.18M were used for MRI measurements. The MRI produces a total amount of fat (g), total lean muscle (g) and total water (g) per mouse. Both the total fat and lean were normalized to the total body weight (g) of the mouse.
Heart weights
Weight-matched mice were used for heart weight analyses for young mice and when possible for aged mice. The body cavity was opened, hearts were removed from the body cavity and blood was drained from the chambers. The heart was then placed on a scale and the weight was recorded. Heart weights were normalized to body weights.
Heart pictures
For heart pictures, the body cavity was opened, hearts were removed from the body cavity, blood was drained from the chambers and a picture was taken of the heart with a reference ruler using an iPhone SE 2020 or a rose gold iPhone 11 Promax. For presentation purposes, heart images were scaled, cropped and placed on a uniform background using Photopea as follows: using the reference ruler, each image was scaled to the same size. Hearts were then cropped using a polygonal lasso select tool with 1 pixel feathering and placed on top of an aquamarine background. A 3-mm scale bar is provided in each figure based on the reference ruler in the images. Uncropped images are included in Supplementary Fig. 1.
Histology
Heart, spleen, lungs, liver and kidneys were harvested and fixed in 10% neutral buffered formalin. Lungs were inflated by injecting the lobes with 1× PBS before fixation. Then samples were routinely processed, paraffin embedded, sectioned at 4–5 μm and haematoxylin and eosin stained. Tissues were evaluated by a board certified veterinary pathologist who was blinded to mouse genotype, age and experimental manipulation, and scored semi-quantitatively for the following parameters: oedema (defined as separation of cardiomyocytes by clear to pale eosinophilic material); heart congestion or haemorrhage (defined as blood vessels expanded by erythrocytes in the myocardium and/or extravascular red blood cells); changes to the cardiomyocytes of the myocardium including pallor and/or cytoplasmic vacuolation, disorganization and/or hypereosinophilia with or without increased size, loss of nuclear detail or loss of cross striations; increased leukocytes within the vessels and parenchyma of the heart; spleen congestion (expansion of the red pulp sinuses by erythrocytes); liver congestion or haemorrhage (expanded sinusoids and/or extravascular red blood cells); increased leukocytes within the sinusoids and parenchyma of the liver; congestion and leucocyte infiltration of the lung interstitium; and kidney congestion (expanded vascular spaces in the cortex and medulla). These parameters were scored on a scale of 0–4 with 0 representing normal tissue; 1 representing minimal changes; 2 representing mild changes; 3 representing moderate changes and 4 representing severe changes relative to a score of 1. Representative images were obtained from glass slides using NIS-Elements BR 3.2 64-bit and plated in Adobe Photoshop CC 2015 and/or Adobe Photoshop 2019. Image white balance, lighting and/or contrast was adjusted using corrections applied to the entire image.
Quantification of E. coli O21:H+ and S. aureus in mouse tissues
For quantification of pathogen in tissues, CFUs were quantified. Spleen, kidney, liver, lung and heart were harvested and homogenized in sterile 1× PBS with 1% Triton X-100 using a BeadMill 24 bench-top bead-based homogenizer (Fisher Scientific). Homogenates were serially diluted and plated on LB agar and EMB-AVNM agar and incubated at 37 °C. Colonies were quantified the following day. The limit of detection for the liver was 50 CFU and 100 CFU for all other tissues examined. Any sample with values below the limit of detection are indicated on the plots as ‘X BLD’ where X indicates the number of mice that had values below the limit of detection for that tissue. Data are plotted as geometric mean ± standard deviation as indicated in the figure legends.
Thermal neutrality experiments
The experiment was performed in a temperature-controlled housing unit (thermal cabinet), purchased from Columbus instruments (model no. ENC52). Mice were housed in the thermal cabinet set to 30 °C, 4 days before infection. At the time of infection, mice were fasted and food was given back 24 h later. Mouse weight, temperature and morbidity were measured every 2 h during the first day of infection. Mice were tracked for survival on subsequent days. A set of control mice were infected at room temperature with the same dose, fasted for 24 h, measured every 2 h during the first day and tracked for survival on subsequent days.
FoxO1 inhibitor infection model
The FoxO1 inhibitor AS1842856 (End Millipore) was injected into mice intraperitoneally at a dose of 40–60 mg kg−1 body weight 36 h before infection and immediately after infection on the opposite side of bacterial injection. Control mice were injected with vehicle. Mice were infected, clinically monitored and used for downstream analyses as described throughout the Methods.
MuRF1 inhibitor infection model
Old mice were orally gavaged with 30 mg kg−1 body weight of the MuRF1 inhibitor EMBL (European Molecular Biology Laboratory) CAS 445222-91-3 (Glixx Laboratories) or vehicle as control 12 h before infection. Mice were then infected with polymicrobial sepsis as described above and 2 h postinfection were gavaged with a second dose of the inhibitor or vehicle. Mice were infected, clinically monitored and used for downstream analyses as described throughout methods.
Leg muscle measurements
Mice were euthanized, and the quadricep, tibialis anterior, extensor digitorum longus, soleus and the gastrocnemius were harvested and weighed to determine the mass of each muscle. Muscle weights were then normalized to the body weight and are shown in the figures.
Homeostatic mouse monitoring
For young and old mouse homeostatic mouse data, mice were weighed daily over the course of ten consecutive days for total body weight tracking. Lean mass, fat mass, free water and total water were determined using Echo MRI. For food consumption, mice were single caged for 48 h before food was weighed every 24 h for 2 consecutive days at the same time of day. Food consumption over the 2 days was averaged to report a grams per day average. For rectal temperature, temperature was measured 3 times per day over a consecutive 5-day period at the same time of day using the Digisense Type J/K/T thermocouple meter. The averages of the three time points per mouse per day were reported. For dissections, mice were euthanized, blood was collected by cardiac puncture and serum stored at −80 °C for future analysis as previously described. Liver, kidney, lung, heart and spleen were harvested and weighed to determine the mass of each organ.
Blood pressure
The non-invasive CODA monitor was used to quantify mean blood pressure, systolic blood pressure, and diastolic blood pressure. Mice were placed in a restrainer and warmed on a far infrared warming platform while recording. The average of 10–15 recordings per mouse per parameter was reported.
Troponin ELISA
Serum from mice infected with polymicrobial sepsis was harvested by cardiac puncture. Troponin I amounts were measured using an ultra-sensitive mouse cardiac troponin I enzyme-linked immunosorbent assay (ELISA) kit (Life Diagnostics) according to the manufacturer’s protocol. Plates were read on a VERSAmax microplate reader manufactured by Molecular Devices and data analysis was done using SoftMax Pro v.5.4.
In vivo metabolic measurements
For the glucose tolerance test, mice were fasted overnight (12 h). Tail tips were cut using a razor blade for blood glucose monitoring using the Nova Max Plus glucose monitoring system. Mice were injected intraperitoneally with 2 g kg−1 of glucose (in PBS) based on total body weight. Blood glucose was measured at 0 (before glucose treatment), 15 min, 30 min, 45 min, 60 min, 90 min and 120 min postinjection. Blood glucose was normalized to percent of time 0, and plotted in Prism where the area under the curve was calculated.
Quantification of whole body metabolic parameters
For quantification of VO2, mice were single caged in metabolic cages within a comprehensive laboratory animal monitoring system Oxymax for Windows v.5.64 24 h before metabolic parameter data collection. Mice were left untouched for roughly 3 days for data collection.
qPCR
For hearts, livers, spleens, kidneys and lungs, organs were harvested and then frozen at −80 °C. They were then homogenized into a tissue powder in liquid nitrogen. The powder was used for subsequent RNA extraction, using the Allprep DNA/RNA Mini Kit (Qiagen) as per the manufacturer’s protocol. For leg muscles, the tissues were harvested and then frozen at −80 °C. They were then homogenized into a tissue powder in liquid nitrogen. Ground muscle tissue powder was added to cold screw cap tube with bead in liquid nitrogen and then homogenized with 800 μl of TRIzol LS Reagent (Invitrogen). Then 160 μl of chloroform (TCI) was added and shaken by hand for 15 s and incubated at room temperature for 3 min. The TRIzol/chloroform lysate was transferred to 1.5-ml tubes and spun in a fume hood centrifuge for 15 min at maximum speed (14,000 rpm). The upper aqueous phase was transferred to a new 1.5-ml tube and 400 μl of isopropanol was added and mixed. The samples were held in a −20 °C freezer for a minimum of 2 h. The samples were added to an Allprep RNeasy spin column (Qiagen) and spun at max speed for 1 min, the flow through was discarded. The samples were then processed according to the Allprep RNA (Qiagen) extraction protocol resulting in 60 μl of diluted muscle RNA. Complementary DNA was generated using SuperScript IV Reverse Transcriptase (Invitrogen) as per the manufacturer’s protocol. Two-step quantitative PCR with reverse transcription (RT–qPCR) was performed using a QuantStudio5 Real-Time PCR instrument (Applied Biosystems) and the QuantStudio5 Design and Analysis software v.1.5.0. Primers sequences are listed in Supplementary Table 1 in the Supplementary Information. The annealing temperature used was 60 °C for Trim63, Fbxo32 and Rps17. A temperature of 58 °C was used for Foxo1.
BNP and GAL3 quantification
Blood was collected by cardiac puncture and serum was stored at −80 °C as described above. For analysis, serum was defrosted and BNP and GAL3 were quantified by a BNP (Cusabio) and GAL3 (Invitrogen) ELISA kit. Samples were diluted at 1:2 for BNP, and at 1:100 to 1:300 for GAL3, and the ELISA was run as specified by both manufacturer’s protocols. Plates were read on a VERSAmax microplate reader manufactured by Molecular Devices and data analysis was done using SoftMax Pro v.5.4.
Western blot analysis
Heart tissue powder were homogenized in 700 μl of Tissue Extraction Reagent II supplemented with Protease Inhibitor Cocktail (100:1) using a BeadMill 24 bench-top bead-based homogenizer (Fisher Scientific). Lysates were centrifuged at 4 °C for 15 min at 27,000 rpm and transferred to a new tube. Samples were quantified with a bicinchoninic acid reaction and subjected to western blot analysis of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (14C10) tabbit monoclonal antibody Cell Signaling 21185 (1:1,000), Phospho-FoxOl (Thr24)/FoxO3a (Thr32) antibody Cell Signaling 94645 (1:1,000), Phospho-FoxOI (Ser256) antibody Cell Signaling 9461 (1:1,000), FoxOl (C29H4) rabbit monoclonal antibody Cell Signaling 28805 (1:1,000), anti-rabbit lgG HRP-linked antibody Cell Signaling 70745 (1:3,000). Samples were mixed with 15 μl of a 1:10 mixture of 2-mercaptoethanol to NuPAGE LDS Sample Buffer (4×) (Invitrogen), then incubated at 70 °C for 10 min and sonicated in a water bath for 10 min. Samples were loaded equally in 7% NuPAGE 1.0 mm × 12 well Tris-acetate gels with 4 °C Tris-acetate SDS running buffer (50 ml 20× Tris-acetate SDS in 950 ml deionized H2O) for 60 min at 150 V. Gels were placed on Trans-Blot Turbo Midi 0.2 µm Nitrocellulose Transfer Packs (Bio-Rad) and inserted into Trans-Blot Turbo Transfer System (Bio-Rad) for 10 min. Membranes were stained with Ponceau Total Protein Stain (Prometheus) to be marked, washed with 1× TBST to destain and placed into blocking solution (5% BSA in 1× TBST) on a shaker platform overnight at 4 °C. Blocking solution was removed and primary antibody solutions were added to membranes (1:1,000 antibody in Prometheus OneBlock Western-CL Blocking Buffer) and placed on a shaker platform overnight at 4 °C. Primary antibody solutions were removed and membranes were washed on a shaker platform for 5 min with 1× TBST, which was repeated 4 times at room temperature. Secondary antibody solution was added containing 1:3,000 anti-rabbit IgG HRP-linked antibody in blocking buffer (5% BSA in 1× TBST) and membranes were placed on a shaker platform for 1 h at room temperature. Secondary antibody solutions were removed and membranes were washed again on a shaker platform for 5 min with 1× TBST, which was repeated 4 times at room temperature. Nitrocellulose blots were developed using a mixture of Femto and Dura chemiluminescent reaction and visualized with a Bio-Rad Gel Doc XR+ Gel Documentation System and Image Lab software v.5.2.1. For total FoxO1 and pFoxO1 blots, the same lysate was run on two different gels at the same time in the same gel rig. The gels were then transferred to the same membrane. After staining of the membrane with Ponceau, the membranes were cut and destained. The cut membranes were then probed for the relevant protein. pFoxO1 and total FoxO1 blots had their own GAPDH loading control that was run on the same gel. All uncropped blots are provided in Supplementary Fig. 1. All primary antibodies were obtained from Cell Signaling Technology and have been validated by the manufacturer for the indicated applications and species. Specifically, the following antibodies are validated for immunoblotting in mouse and rat samples according to the manufacturer’s website: GAPDH (14C10) rabbit monoclonal antibody (no. 21185) validated for western blot, mouse, rat and human. Phospho-FoxO1 (Thr24/FoxO3a (Thr32) antibody (no. 94645) validated for western blot, mouse, rat and human. Phospho-FoxO1 (Ser256) antibody (no. 9461) validated for western blot, mouse, rat and human. FoxO1 (C29H4) rabbit monoclonal antibody (no. 28805) validated for western blot, mouse, rat and human. Anti-rabbit IgG, HRP-linked antibody (no. 70745) validated for use as a secondary antibody in western blot applications. All antibodies were used at the dilutions recommended by the manufacturer and produced bands at the expected molecular weights in our samples, consistent with the manufacturer’s validation data.
Serum analysis
Sera harvested by cardiac puncture were analysed by IDEXX Bioanalytics for total bicarbonate, potassium, anion gap, cytokines, AST, ALT, BUN, albumin and creatinine.
RNA-seq and data processing
Total RNA was extracted from heart tissue using the Allprep DNA/RNA Mini Kit (Qiagen) per manufacturer protocol. Libraries were generated using the Illumina TruSeq Stranded messenger RNA Sample Prep Kit following the manufacturer’s instructions (Illumina). Then 75-base pair single-end sequencing was preformed using the Illumina HiSeq 2500 platform. Read quality was assessed using FastQC, version 0.11.5 (Babraham Bioinformatics). Reads were mapped to the mm10 genome using STAR v.2.5.3a (ref. 44). Gene expression levels were quantified across all exons using HOMER v.4.10 (ref. 45). Differential gene expression analysis was carried out by using edgeR v.3.26.7 (refs. 46,47). Results corrected for multiple hypotheses testing using the Benjamini–Hochberg method48. The false discovery date threshold for significance was set at 0.05 or lower and a log2 fold-change of 1 or greater. Heat maps and clustering were performed using R v.3.6.1 (R Core Team) using variance stabilizing transformed counts from DESeq2 v.1.24.0. Graphical packages (pheatmap v.1.0.12, ggplot2 v.3.3.2, gplots v.3.0.1.1, RColorBrewer v.1.1-2, VennDiagram v.1.6.20) were used to visualize data by hierarchical clustering, and generating heat maps, Venn diagrams and principal components analysis plots. Gene Ontology and KEGG analyses were done using DAVID49,50.
Statistics and data presentation
GraphPad Prism 10 for Mac OS X version 10.1.1 and Microsoft Excel v.16.78 were used for graphing of data and statistical analyses. For survival, log-rank analysis was used. D’Agostino and Shapiro–Wilk normality tests were performed on data sets to determine the distribution of the data sets. For pairwise comparisons, unpaired t-test, Mann–Whitney test, one-way analysis of variance (ANOVA) or Kruskal–Wallis test with Tukey or Dunn’s test, or two-way ANOVA with Tukey or Sidak’s multiple comparison were performed. Statistical tests used are noted in the figure legends. In accordance with our IACUC guidelines, in an effort to reduce the number of animals used for our experiments, where possible and appropriate, experiments were done in parallel and shared controls were used. This is indicated in the figure legends where relevant. For some experiments, data are shown in several panels to simplify viewing of different comparisons within the data sets. This is indicated in the figure legends where relevant. Power analyses showed the appropriate sample sizes to use for 80% power to detect an estimated detectable effect size, assuming a 5% significance level and a two-sided test. Allocation was random within sex-matched and age-matched cohorts. For data exclusions, see Fig. 1 for the troponin I measurements. We had to estimate the appropriate amount to dilute samples for the ELISA. Four uninfected young serum samples and one young surviving infected serum sample were diluted below the limit of detection. We did not have enough serum for these samples to rerun at a greater concentration, so we omitted these samples from the analysis. In Extended Data Fig. 4b–f,h–j, a pathologist flagged one young surviving and three young dying slides as having artefacts present that can compromise interpretations and one young surviving and two young dying slides as having severe artefacts present and slides of poor quality, therefore, these were omitted from data plots. In Extended Data Fig. 7h a pathologist flagged one lung from a vehicle surviving mouse as a possible artefact. This sample was omitted. In Extended Data Fig. 8i–p, a pathologist flagged one wild-type infected slide as having artefacts, and it was not scored. We still had sufficient sample sizes even with these samples omitted. Experiments were performed with biologically independent animals for each condition and experiments were repeated independently as indicated in the figure legends. Allocation was random within sex-matched and age-matched cohorts. Within each genotype, mice of the same sex and age were randomly assigned to experimental groups. Littermates were used whenever possible. Independent experimental replicates included mice from different litters and cages and were performed on different days, allowing potential litter, cage or day effects to be identified. Potential confounding factors were controlled through experimental design and replication. Blinding was not possible for animal infection experiments because disease outcomes (for example, survival and visible morbidity) were readily apparent to investigators. For quantitative assays including ELISAs, qPCR, western blots, blood chemistry, histopathology, tissue weights, the individuals performing the assays and/or analyses were blinded to group allocation. Data for ELISAs, qPCRs and blood chemistry were collected by machine-based, unbiased, quantitative means. A single representative experiment or many independent experiments combined are shown in the figures. This is noted in the figure legends.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.