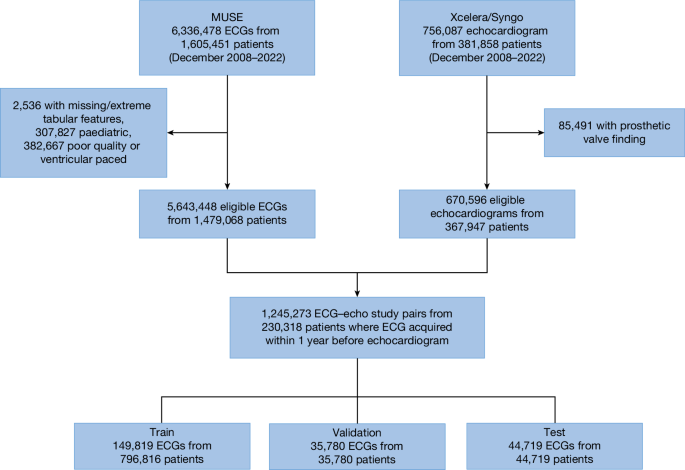
Patient identification and data sources
Patients 18 years of age or older who underwent a digitally-stored 12-lead ECG between December 2008 and 2022 at one of eight NYP-affiliated hospitals (Columbia University Irving Medical Center, Weill Cornell Medical Center, NYP-Brooklyn Methodist Hospital, NYP-Lower Manhattan Hospital, NYP-Queens Hospital, NYP-Allen Hospital, NYP-Westchester Hospital and adult patients at the Morgan Stanley Children’s Hospital of New York) were identified. Those 230,318 unique patients who had a diagnostic quality, non-ventricularly paced ECG performed up to 1 year before an echocardiogram formed the NYP multicentre cohort, with 1,245,273 distinct ECG–echocardiogram pairs (Fig. 1). Demographics, including race and ethnicity, were abstracted from the electronic medical record.
ECG data were accessed from the MUSE data management system (GE Healthcare) at each institution. ECG data that were abstracted included demographic and ECG-specific tabular information, including age, sex, atrial and ventricular rates, and pulmonary regurgitation, QRS and Bazett’s corrected Q wave-to-T wave intervals. The ECG waveform data were abstracted at 250 Hz for all 12 ECG leads for a total of 30,000 data points.
Echocardiographic data were accessed from the Syngo Dynamics (Siemens Healthineers) and Xcelera (Phillips) systems. Abstracted data included the LVEF, interventricular septum and posterior wall thicknesses (with the larger being defined as the maximum low left ventricular wall thickness), qualitative right ventricular systolic function (defined as normal, mildly reduced, moderately reduced or severely reduced), the PASP and maximum tricuspid regurgitation maximum velocity, the presence of a pericardial effusion (normalized to a scale of none or trace, small, moderate or large) and the severity of the VHDs of aortic stenosis, aortic regurgitation, mitral regurgitation, tricuspid regurgitation and pulmonic regurgitation (normalized to a scale of none or trace, mild, moderate or severe). Mild to moderate VHD was classified as mild disease and moderate to severe VHD was classified as moderate disease. Repaired or replaced heart valves were excluded from the dataset. These data were harmonized across each echocardiographic reading system and hospital with a minimum of 100 cases audited per label to confirm the accuracy of each analysis. From these features, we defined the presence or absence of SHD for each echocardiogram using the following binary cutoffs: LVEF (less than or equal to 45%), maximum low left ventricular wall thickness (greater than or equal to 1.3 cm), right ventricular dysfunction (moderate or severely reduced), pulmonary hypertension (PASP greater than or equal to 45 mm Hg or tricuspid regurgitation jet velocity greater than or equal to 3.2 m s−1), aortic stenosis (moderate or severe), aortic regurgitation (moderate or severe), mitral regurgitation (moderate or severe), tricuspid regurgitation (moderate or severe), pulmonary regurgitation (moderate or severe) and a significant pericardial effusion (moderate or large). For an ECG to be labelled as being ‘positive’ for a disease, it must have been performed within 1 year before an echocardiogram with SHD. In patients without SHD (confirmed by at least one ‘negative’ echocardiogram), all ECGs before the most recent echo were labelled as negative and included in the study. Only ECGs with an echo occurring afterwards were used to ensure no ECGs occurring after corrective procedures in which a future echo may not occur were included as they would be mislabelled.
To be included in the study, an echocardiography report was required to include LVEF, a wall thickness measurement and one relevant valve finding. Missing data were imputed using the following process. For valve findings, if either regurgitation or stenosis was commented on, the other was presumed normal (for example, if aortic stenosis was reported but aortic regurgitation was not, then we assumed no aortic regurgitation). If not specifically commented on, a pericardial effusion and pulmonary hypertension were presumed to be absent.
In addition to this base SHD label, a secondary, more stringent cutoff was defined for each endpoint (for example, LVEF less than or equal to 35%) to reflect ‘severe SHD’. These cutoffs and model accuracy using these severe SHD endpoints are detailed in the Supplementary Information.
For the primary analysis, data from all eight hospital campuses were blended and split by patient into training, validation and test sets (64%, 16% and 20%). Further experiments are detailed in the Supplementary Information; for example, using alternate data partitions to hold out specific NYP hospitals from training to assess generalization. In all cases, several ECG–echocardiogram pairs were used in training, but the most recent ECG–echocardiogram pair was selected for each unique patient in the validation and test sets. This retrospective study was conducted with approval of the Columbia University and Weill Cornell Institutional Research Boards with waiver of patient consent.
Model details
The EchoNext model comprises a convolutional neural network that takes a digital 12-lead ECG waveform, patient demographics and ECG-specific tabular information to predict the presence or absence of SHD (Supplementary Table 1). Extending previous work3, we trained EchoNext as a multitask classifier such that separate terminal branches of the model predict the presence of the SHD composite label and the presence of an individual component label (for instance, the presence or absence of aortic stenosis), respectively. Details on the model design, hyperparameters, testing and optimization are addressed in the Supplementary Information.
Silent deployment validation
As the model development dataset comprised ECGs acquired throughout December 2022, we subsequently collected ECG–echocardiogram pairs acquired at NYP from January to 16 September 2023, as a temporally distinct validation set. Patients included in the development cohort were excluded from this analysis.
Prospective validation of ValveNet and EchoNext
Before development of EchoNext, study investigators had created ValveNet, a similarly architected AI-ECG model trained to detect the left-sided VHD of aortic stenosis, aortic regurgitation and mitral regurgitation, a subset of SDH3. To test the ability of a system using this model to detect clinically significant cardiac disease, we designed the Aortic Stenosis Discovery Study, a 100-patient, open-label trial. Adult patients were eligible if they had a digital 12-lead ECG performed at Columbia University and had no history of an echocardiogram within the last 3 years in our system, no history of left-sided VHD and no dementia or other non-cardiac life-limiting disease with expected survival less than 1 year. Eligible patients were recruited by their ValveNet score (a continuous variable from 0–1 with a value closer to 1 indicating a higher model confidence that VHD was present) into high-risk (score greater than or equal to 0.6) or moderate-risk (score 0.3–0.6) groups. Patients with scores less than 0.3 were excluded due to a very low predicted risk of cardiac disease. Consented patients underwent an echocardiogram. The primary endpoint was moderate or severe aortic stenosis, aortic regurgitation or mitral regurgitation. The key secondary endpoint was any SHD that was identical to the EchoNext label. Critically important findings were communicated with patients and physicians, and appropriate clinical follow-up was coordinated by study investigators for newly diagnosed disease.
Cardiologist survey
Board-certified attending cardiologists were recruited from Columbia University to study human accuracy in the detection of SHD using the ECG. A total of 13 cardiologists were recruited to take this study (J.M.D., S.Y., G.F.R., S.R.A., Q.L., C.K.B., P.V., C.A.W., E.M.D., V.A., M. Lebehn, P.N.K. and S.S.). A total of 150 ECGs were selected from the NYP multicentre test set representing a similar age distribution and SHD prevalence to the entire dataset. The digital ECG was accessed as a pdf and the name, date and clinical interpretation was cropped out of the image leaving only the waveform and the ECG measurements (ventricular rate, pulmonary regurgitation interval, QRS interval, Q wave-to-T wave interval and axis). The age (truncated to greater than 90) and sex were added to each ECG. These 150 ECGs were split into blocks of 50. Each cardiologist was presented with a block of 50 ECGs and were asked to answer two questions for each ECG: whether the patient was likely to have SHD (not likely or likely). After completion of each block of 50 ECGs, they were given the same 50 ECGs with the addition of the AI model analysis with both the model output (0–1) and model interpretation (less than 0.6 not consistent with SHD, greater than or equal to 0.6 consistent with SHD) added to the image. Each cardiologist could complete up to 300 ECGs (150 without and 150 with the AI model analyses). The results from all the cardiologists were pooled for primary analysis with calculation of the accuracy, sensitivity and specificity using standard methods with 95% CIs for accuracy calculated by the Clopper–Pearson method. The accuracy of the EchoNext model in this 150 ECG dataset was determined using a threshold of 0.6. This method of human–machine comparison is similar to typical methods and one we have used previously34,35. Clinically normal ECGs were identified from the clinical interpretation report and performance was compared between normal and abnormal ECGs.
Statistical analysis
Descriptive statistics were used to describe the data using standard methods. The performance of EchoNext was assessed using standard metrics, including AUROC and AUPRC. Diagnostic odds ratio was also computed at the operating point of 0.5. For each statistical test, 95% CIs were generated using 1,000 bootstrapped estimates. Subgroup analyses were performed using subsets of age, sex, race and ethnicity. All statistical analyses were performed using Python v.3.8.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.