Insects
Gregarious and solitary locusts (L. migratoria) used in the experiments were maintained at the Institute of Zoology, Chinese Academy of Sciences, Beijing, China. In brief, gregarious locusts were reared in cages (30 cm × 30 cm × 30 cm) with 800 to 1,000 first-instar insects per cage in a well-ventilated room. Solitary locusts were raised in another room, each in a separate ventilated cage (10 cm × 10 cm × 25 cm). Gregarious and solitary locusts were maintained for at least three generations before the experiments. All locusts were cultured under the following conditions: photoperiod light 14 h:dark 10 h, temperature 30 ± 2 °C, relative humidity 60 ± 5%, and a diet of fresh greenhouse-grown wheat seedlings and bran.
Volatile collection and 4VA quantification
The volatiles of fifth-instar gregarious and solitary nymphs were collected by static solid phase microextraction (SPME) and detected by a Bruker GC system (456-GC) coupled with a triple-quadrupole mass spectrometer, as described1. In brief, a fibre (PDMS/DVB 65 μm, 57310-U) was introduced into a glass jar (10.5 cm high × 8.5 cm internal diameter) approximately 1 cm above a stainless steel lid (9 cm in diameter with holes of 2 mm diameter and 2 mm apart), which served as a barrier to confine a group of five fifth-instar locusts. The SPME volatiles collected from an empty glass jar for 30 min served as a control. The fibres with adsorbed odours were subjected to chemical analyses.
Starvation of locusts
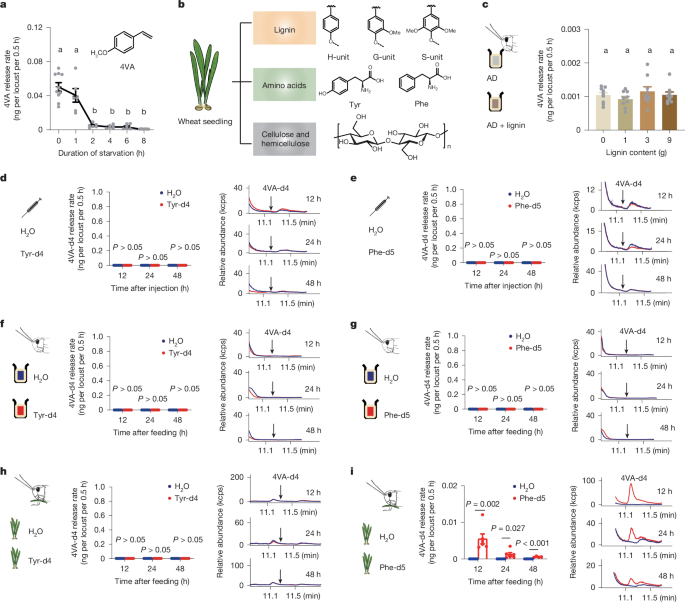
Fifth-instar gregarious nymphs were cultured in perspex cages (15 cm × 15 cm × 15 cm) for starvation treatment. The locusts were first fed with sufficient wheat seedlings, and the starvation time of the locusts was 1 h, 2 h, 4 h, 6 h and 8 h. After treatments, the volatiles of locusts were collected using SPME, and gas chromatography–mass spectrometry (GC–MS) analysis was used to quantify 4VA emissions. The experiment consisted of six biological repetitions with four locusts per repetition. A Bruker chemical analysis MS workstation (v.8.0) was used to analyse and process the data.
Injection of deuterated compounds
Phe-d5, Tyr-d4, PAH-d6, CA-d6, p-HCA-d4 and 4VP-d4 were dissolved with sterile water at the concentration of 10 μg μl−1; the deuterated compounds (2 μl) were injected into the abdomen cavity of early fifth-instar gregarious nymphs. The control group was injected with 2 μl sterile water. The volatiles of locusts were collected by SPME and quantified by GC–MS at 12 h, 24 h and 48 h after injection.
Feeding of lignin with an artificial diet
Locust artificial diet was made according to Supplementary Tables 2 and 3. In brief, 400 ml ultra-pure water and all compounds without vitamin C and vitamin B1 were thoroughly mixed and then placed in a high-temperature sterilizer for 20 min to fully dissolve the agar powder. The dissolved solution was placed at room temperature to 30–40 °C; vitamin C and vitamin B1 were added and the solution was stirred thoroughly until uniform. One, three and nine grams of lignin powder (Sigma-Aldrich, 370959) was added to 50 ml of artificial diet before being cooled and coagulated. The early fifth-instar gregarious nymphs were selected and starved for 6–8 h. The locusts in the experimental group were fed an artificial diet containing lignin, while the control group locusts were fed an artificial diet without lignin. A fresh artificial diet was applied once per 12 h, and the locusts were fed for 24 h. SPME was used to collect volatiles of locusts, and GC–MS was used to quantify the release of 4VA.
Feeding of deuterated compounds with artificial diet or plants
Phe-d5, Tyr-d4, PAH-d6 and CA-d6 were dissolved in sterile water at 10 μg μl−1. Phe-d5, Tyr-d4, PAH-d6 and CA-d6 solutions were mixed with an artificial diet or sprayed uniformly on the stems and leaves of wheat seedlings. The fifth-instar gregarious nymphs were fed with artificial diet or wheat seedlings supplemented with deuterated compounds. The control group was fed with an artificial diet of wheat seedlings sprayed with sterile water. The volatiles of gregarious locusts were collected after 12 h, 24 h and 48 h of feeding, respectively, and GC–MS was used to detect 4VA release by the locusts.
Chemical analysis
A Bruker GC system (456-GC) coupled with a triple-quadrupole mass spectrometer (Scion TQ MS/MS, Bruker Daltonics) equipped with a DB-1MS column (30 m × 0.25 mm i.d. × 0.25 μm film thickness, Agilent Technologies) was used to quantify the volatile compounds in the SPME samples as described in the previous studies1,14. A Bruker chemical analysis MS workstation (MS Data Review, Data Process, v.8.0) was used to analyse and process the data. Standard compounds (purities ≥ 95%, Sigma-Aldrich) with different dosages (0.1, 1, 10, and 100 ng μl−1) were used to develop the standard curves to quantify the volatiles. The same thermal program and MRM method were used.
The volatile profiles of locusts after RNAi knockdown of 4VPMTs and feeding of 4NP were quantified using an Agilent GC system (5973N) coupled with a mass spectrometry system equipped with a HP-5MS column (60 m × 0.25 mm i.d. × 0.25 μm film thickness, Agilent Technologies) to quantify the volatile compounds in the SPME samples. The initial temperature of the oven was maintained at 40 °C for 1 min, and the ramp was at 20 °C min−1 to 300 °C (hold 2 min). The injector temperature was maintained at 250 °C with a constant flow rate of 1.0 ml min−1 of helium. The GC–MS electron impact source was operated in full scan mode with the MS source temperature at 240 °C and MS Quad at 150 °C. Volatile compounds were identified by comparing their retention times with the synthetic standards in the same column. The referenced mass spectra were from the NIST11 library (Scientific Instrument Services).
Derivatization and quantification of biosynthetic intermediates
Phenylalanine, cinnamic acid, p-HCA and 4VP in guts, haemolymph and legs of gregarious and solitary locusts were derivatized and quantified by GC–MS/MS. Approximately 100 mg of fresh tissue samples were weighed and transferred into a centrifuge tube containing 500 μl of extraction reagent (ethanol: acetonitrile, 9:1) and homogenized thoroughly in a grinder (JXFSTRP-32L, Shanghai Jingxin Industrial Development, 60 Hz, 2 min). After stewing for 15 min at room temperature, the samples were centrifuged at 12,000 rpm for 10 min at 4 °C. The supernatants were transferred to new centrifuge tubes for vacuum drying. Methoxyaminutese hydrochloride (20 mg ml−1, dissolved in pyridine, 50 μl each sample) was mixed thoroughly in the dried samples, and the samples were kept at 37 °C for 90 min. N-Methyl-N-(trimethylsilyl) trifluoroacetamide (70 μl each sample) was subsequently added to the samples, and the samples were placed at 37 °C for 90 min. The samples were centrifuged at 12,000 rpm for 10 min at 4 °C. The supernatants were stored at −20 °C for detection.
RNA sequencing and data processing
Total RNA was extracted from hind legs (3 samples with 5–6 individuals per sample) using Trizol reagent following the manufacturer’s instructions. The quantity and purity of the total RNA were determined in an Agilent 2100 Bioanalyzer (Agilent) to verify RNA integrity. After the QC procedure, the RNA with poly-A in eukaryotic total RNA was enriched by TIANSeq mRNA Capture Kit (TIANGEN). Then, using the captured RNA as the starting sample, the TIANSeq Fast RNA Library Kit (Illumina) was used to construct the transcriptome sequencing libraries. In brief, the transcriptome sequencing library was built through RNA random fragmentation, cDNA strand 1 and strand 2 synthesis, end repair, A-tailing, ligation of sequencing adapters, size selection and library PCR enrichment. Library concentration was first quantified using a Qubit 2.0 fluorometer (Life Technologies) and then diluted to 1 ng μl−1 before checking insert size on an Agilent 2100 and quantifying to greater accuracy by qPCR (library activity > 2 nM). The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumina) according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on an Illumina sequencing platform and 150 bp paired-end reads were generated. The raw reads of 6 samples are available for download from the NCBI Sequence Read Archive (SRA) server (accession number: PRJNA1046724).
RNA preparation and qPCR assay for genes
Eight samples of locust hind legs (2–3 individuals per sample) were collected and homogenized in Trizol reagent (Life Technology) and the total RNA was extracted following the manufacturer’s instructions. DNase was applied to eliminate DNA contamination in the RNA samples. We reverse transcribed 2 µg of total RNA in every sample using MMLV reverse transcriptase (Promega) by the manufacturers’ instructions to analyse the expression levels of mRNAs. PCR amplification was conducted with the Roche Light Cycler 480 using a miRcute miRNA qPCR Detection Kit (Tiangen) and a Real Master-Mix (SYBR Green) kit (Tiangen), respectively. RP49 was used as an endogenous control for mRNAs. The amplification procedure followed the manufacturers’ protocols, and the melting curve was detected to confirm the amplification specificity of the target genes. All PCR amplifications are sequenced to verify the specificity of primers. The primer sequences for the qPCR assay are provided in Supplementary Table 4.
RNAi and GC analysis
After designing the fragments of LOCMI16699, LOCMI02868, LOCMI17143, LOCMI17606, LOCMI16705 and LOCMI03758 sequences for RNAi, we blasted the LOCMI16699 fragments against the LOCMI02868 sequences of the migratory locust to detect sequence homologies. We selected the non-homologous fragment with other genes in the genome database to avoid non-specificity during RNAi knockdown. dsRNAs of GFP, LOCMI16699, LOCMI02868, LOCMI17143, LOCMI17606, LOCMI16705 and LOCMI03758 were prepared using the T7 RiboMAX Express RNAi System (Promega). Fifth-instar gregarious locusts were selected for injection. Ten micrograms of double-stranded RNAs of LOCMI16699, LOCMI02868, LOCMI17143, LOCMI17606, LOCMI16705, LOCMI03758 and double-stranded RNAs of GFP (as a control), respectively, were injected into the second abdominal segment of gregarious locusts. The effect of RNAi on relative mRNA and 4VA production levels was investigated by qPCR and GC–MS/MS after injection for 72 h. The primers for RNAi are provided in Supplementary Table 5.
Western blot
Hind leg samples from six groups of crowded solitary locusts and isolated gregarious locusts were collected and homogenized in TRIzol reagent (Life Technology), and protein for western blot analysis was extracted following manufacturers’ instructions, respectively. Custom-made affinity-purified polyclonal antibodies against 4VPMT1 (mouse) (ABclonal), 4VPMT2 (rabbit) (ABclonal) and GAPDH19 were used for protein analyses. The specificity of the two antibodies were verified by western blot after the knockdown of 4VPMT1 and 4VPMT2, respectively (Supplementary Fig. 1a,b). The protein samples (10 μg μl−1) were separated by gel electrophoresis and then transferred onto polyvinylidene difluoride membranes (Millipore). Non-specific binding sites on the membranes were blocked with 5% skim milk. The blots were separately incubated with the primary antibody (mouse anti-4VPMT1, 1:5,000; rabbit anti-4VPMT2, 1:1,000; rabbit anti-GAPDH, 1:5,000) in 5% skim milk overnight at 4 °C. After incubation; the membranes were washed by PBST (PBS with 0.1% Tween), then incubated with anti-rabbit IgG secondary antibody (1:5,000) (EASYBIO Technology) for 1 h at room temperature, and then washed three times. An ECL kit subsequently detected the immunological blot (Thermo Fisher, 34096). The intensities of the western blot signals were quantified using ImageJ (v.1.51k).
Immunofluorescence of 4VPMT1 and 4VPMT2 in the locust hind leg
The dissected hind legs were embedded in freezing medium Tissue-Tek O.C.T. Compound (Sakura Finetek) and rapidly frozen at –70 °C. Sections (10 μm) were prepared at –20 °C (Leica CM1950), thaw mounted on SuperFrost Plus slides (Menzel-Gläser) and air dried for 15 min. The immunofluorescence was performed according to the protocol described in a previous study20 with slight modifications. After fixed in 4% formaldehyde at room temperature for 1 h, the sections were washed with 0.1 M PBS (pH 7.2) twice for 15 min each, and then incubated in 0.1 M PBS containing 5% normal goat serum (NGS, Boster) for 1 h at room temperature. The primary anti-4VPMT1 and 4VPMT2 antibody (custom made, ABclonal) was diluted at 1:1000 and 1:500 in 0.1 M PBS containing 2% NGS, respectively. Incubation with primary antibodies lasted for 24 h. The tissues were washed with 0.1 M PBS three times for 15 min each and subsequently incubated with the secondary antibodies, goat anti-rabbit antibody Alexa fluor 488 (1:500, Life Technology, A11034) and goat anti-mouse antibody Alexa fluor 546 (1:500, Life Technology, A11030) for 1 h at room temperature. After washing three times, the tissues were mounted in anti-fade fluorescence mounting medium. The negative serum was used as the negative control. The nucleus of locust hind leg was labelled by Hoechst33342 (Life Technology). Confocal images were obtained by a Zeiss LSM 710 confocal microscope (Zeiss) equipped with ZEN 2012 software.
Protein expression and purification
The DNA fragments encoding 4VPMT1 and 4VPMT2 were amplified by PCR using the primers pET28a-4VPMT1-F/R and pET28a-4VPMT2-F/R (Supplementary Table 6), respectively, and cloned into a pET28a-derived vector. The constructs with an N-terminal His tag were transformed into the E. coli BL21 (DE3) strain, and the bacteria were cultured in LB medium at 37 °C. When the OD600 of the culture reached 0.5, the expression of 4VPMT1 and 4VPMT2 was induced by the addition of 0.4 mM isopropyl-β-d-thiogalactopyranoside at 16 °C for 14 h. The cells were collected by centrifugation at 4,500g for 15 min and then resuspended in a binding buffer (30 mM imidazole, 30 mM Tris-HCl, pH 8.0, 50 mM NaCl). The supernatants were transferred into new centrifuge tubes after ultrasonication and centrifugation at 15,000g for 15 min. Ni2 + -NTA agarose resin was first equilibrated with 30 mM imidazole buffer. After adding supernatant, the target protein was washed with 50 mM imidazole buffer and then eluted with 250 mM imidazole buffer. The purity of the eluted protein was estimated by SDS–PAGE, and the protein sample was stored at −80 °C. The mutant proteins were expressed, purified, and stored in the same manner as the wild-type protein.
Enzymatic activity assays
The enzymatic activity of 4VPMTs was measured by quantifying the amount of product in the reactions using SPME headspace, as described above, with some modifications. To determine the kinetic parameters of 4VPMT1 for 4VP, 399 μl of the reaction mixture was pipetted into a sample vial (RY-10100, 5 ml for headspace adsorption) containing buffer (20 mM Na2HPO4-NaH2PO4, 50 mM NaCl, pH 7.5), 20 μg 4VPMT1 protein and 4 μl SAM (100 mM). The reaction was started by adding 1 μl substrate 4VP (0.04–100 mM) at various concentrations, and the vial was then sealed with a cap (RY-10100, Sample bottle with clamp cap). The handle rod was held in a horizontal position with the sample vial, then the extraction head was slowly pushed into the sample vial at a distance of 3–4 cm from the bottle mouth. The fibre was pushed out to expose the headspace volatiles generated by the reaction, and the adsorption time was 10 min at 30 °C. To determine the kinetic parameters of 4VPMT2 for 4VP, 399 μl of the reaction mixture was pipetted into a sample vial (RY-10100, 5 ml for headspace adsorption) containing buffer (20 mM Na2HPO4-NaH2PO4, 50 mM NaCl, pH 7.5), 180 μg 4VPMT2 protein and 4 μl SAM (100 mM); the reaction was started by adding 1 μl 4VP (4–320 mM) at various concentrations, and the vial was then sealed with a cap (RY-10100, Sample bottle with clamp cap). The products were collected using SPME for 20 min at 30 °C. The amounts of product in the reactions were calculated according to the standard curve of 4VA. Data fitting was performed using GraphPad Prism 8, and Km, kcat, kcat/Km and ki values represent the mean ± s.e.m. of three independent replicates.
The inhibition assay of 4VPMT1 against 4VP using 4VP analogues was carried out by pipetting 399 μl of the reaction mixture into a sample vial (RY-10100, 5 ml for headspace adsorption) containing buffer (20 mM Na2HPO4-NaH2PO4, 50 mM NaCl, pH 7.5), 20 μg 4VPMT1 protein, 4 μl SAM (100 mM) and 1 μl analogues (4 mM); the reaction was started by adding 1 μl 4VP (0.4 mM), and the vial was then sealed with a cap (RY-10100, Sample bottle with clamp cap). The products were collected using SPME for 10 min at 30 °C and analysed by GC–MS. The amount of product in the reactions with or without 4VP analogues was calculated according to the standard curve of 4VA. Data were analysed by a two-tailed unpaired t-test using GraphPad Prism 8 software.
The IC50 values of 4NP and 4-trifluoromethylphenol to the enzymatic activity of 4VPMT1 were determined exactly as previously described. Specifically, 1 μl of 4NP or 4-trifluoromethylphenol at various concentrations (0.00004–20 mM) was added to the reaction buffer containing 20 mM Na2HPO4-NaH2PO4 (pH 7.5), 50 mM NaCl, 20 μg 4VPMT1 protein and 4 μl SAM (100 mM). The enzymatic reaction was initiated by adding 1 μl of 0.4 mM 4VP, and the adsorption time was 10 min at 30 °C. To determine the IC50 values of 4NP and 4-trifluoromethylphenol to 4VPMT2, 1 μl of 4NP or 4-trifluoromethylphenol at various concentrations (0.00004–20 mM) was added to the reaction buffer containing 20 mM Na2HPO4-NaH2PO4 (pH 7.5), 50 mM NaCl, 180 μg 4VPMT2 protein and 4 μl SAM (100 mM). The enzymatic reaction was initiated by adding 1 μl of 30 mM 4VP, and the adsorption time was 20 min at 30 °C. The IC50 values were calculated by plotting the relative activity against various concentrations of testing compounds with a dose-response-inhibition function in GraphPad Prism 8 software.
To determine the kinetic parameters of 4VPMT1 for 4NP, 399 μl of the reaction mixture was pipetted into a sample vial (RY-10100, 5 ml for headspace adsorption) containing buffer (20 mM Na2HPO4-NaH2PO4, 50 mM NaCl, pH 7.5), 200 μg 4VPMT1 protein and 4 μl SAM (100 mM). The reaction was started by adding 1 μl substrate 4NP (0.04–40 mM) at various concentrations, and the vial was then sealed with a cap (RY-10100, Sample bottle with clamp cap). The products were collected for 30 min at 30 °C. After adsorption, the fibre extraction head was retracted and then the protective needle was withdrawn from the sample vial to prepare for GC–MS analysis, including 3 biological replicates. Different concentrations of 4NA standard sample (1–100 ng μl−1) were configured and analysed using GC–MS. The amounts of product in the reactions were calculated according to this standard curve of 4NA. Data fitting was performed using GraphPad Prism 8, and Km, kcat, and kcat/Km values represent the mean ± s.e.m. of three independent replicates. To determine the kinetic parameters of 4VPMT2 for 4NP, 399 μl of the reaction mixture was pipetted into a sample vial (RY-10100, 5 ml for headspace adsorption) containing buffer (20 mM Na2HPO4-NaH2PO4, 50 mM NaCl, pH 7.5), 1,800 μg 4VPMT2 protein and 4 μl SAM (100 mM), and the reaction was started by adding 1 μl 4NP (0.4–400 mM) at various concentrations, and the vial was then sealed with a cap (RY-10100, Sample bottle with clamp cap). The products were collected using SPME for 40 min at 30 °C. The amounts of product in the reactions were calculated according to the standard curve of 4NA. Data fitting was performed using GraphPad Prism 8, and Km, kcat, kcat/Km and ki values represent the mean ± s.e.m. of three independent replicates.
Crystallization of 4VPMT2, data collection and structure determination
The DNA fragment encoding 4VPMT2 was amplified by PCR from the pET28a-4VPMT2 plasmid using the primers pQLinkH-4VPMT2-F/R (Supplementary Table 6) and cloned into a pQlinkH vector. The construct with an N-terminal His tag was transformed into the E. coli MC1061 strain. The overexpression and purification of 4VPMT2 in the MC1061 strain followed the same protocol as the BL21 (DE3) strain described above. To prepare the protein sample for crystallization, N-His-4VPMT2 was further purified by the size exclusion chromatography using a Superdex increase 200 10/300 GL column (GE Healthcare) with buffer (30 mM Tris, 200 mM NaCl, pH 8.0). The peak fraction was collected and concentrated to 10 mg ml−1. Crystals were grown using a sitting drop vapour diffusion method at 18 °C. Pre-incubated 4VPMT2 with SAM was crystallized in 0.1 M MMT buffer (malic acid, MES and Tris buffers), pH 7.0, and 25% PEG1500. The crystals were soaked for about 24 h in the respective reservoir solution with a final concentration of 4VP of 2.5 mM. Crystals were cryoprotected in a reservoir solution supplied with 20% ethylene glycol and flash-frozen immediately in liquid nitrogen. X-ray diffraction data were collected at the Shanghai Synchrotron Radiation Facility (beamline BL02U1) with an X-ray wavelength of 1.0 Å at a temperature of 100 K. The data were processed using HKL2000 (HKL Research). The structure was solved by a molecular replacement method using Phaser, with the predicted structure from AlphaFold v2.0 (refs. 21,22) as the search model and refined using Coot and Phenix.
3D structure prediction of 4VPMT1
The full-length amino acid sequence of 4VPMT1 was used for structure prediction by AlphaFold v2.0. The monomeric structure of 4VPMT1 was predicted. Parameters -m is model 1, model 2, model 3, model 4, model 5, -t is 2022-08-21, and -g False. The models with the highest confidence level ‘ranked_0.pdb’ were selected as the predicted structure for 4VPMT1. The predicted structures share a backbone similar to the resolved structures of homologous JHAMT (PDB ID: 7EBS, 7EBX and 7V2S) and 4VPMT2 (8ZSA), indicating high predictive accuracy. The per-residue confidence score (pLDDT) of amino acids 1 to 54 of 4VPMT1 is below 70, and thus this region was removed in further structural analysis.
Docking calculations
As 4VPMT1 shares analogous structural backbones to 4VPMT2, the positions of SAM and 4VP in 4VPMT1 were constructed according to their locations in 4VPMT2 (8ZSA). Similarly, the substrate competitor 4NP was docked into the pocket of 4VPMTs according to the position of the substrate 4VP. Molecular docking was accomplished by Schrödinger 2021-2. Preparation of the ligand was carried out via the default parameters of ligPrep. Protein preparation was carried out by Protein Preparation Wizard, with the force field as OPLS_2005 in restrained minimization23. According to the reaction mechanism, the cofactor SAM is involved in the reaction process, so the docking pocket for the protein was chosen as a 20 Å × 20 Å × 20 Å size with SAM as the centre of the grid box. There is no constraint on small molecules.
Molecular dynamics simulations
The complexes of 4VPMTs with their ligands obtained from the docking calculations were used to perform molecular dynamics simulations. Molecular dynamics simulations were performed in desmond (Schrödinger). The solvent was predefined using the TIP3P parameters. The solution simulated was a 0.15 M NaCl solution. NaCl was used to neutralize the charge of the system. The simulations were carried out at 300 K. The simulations were performed using Desmond’s standard NPT relaxation protocol. No restrictions are added to the ligands during this process. A total of three sets of 500 ns computational simulations were carried out. The results of the simulations were subsequently analysed for protein–ligand interaction, from which information on PL-RMSD and PL-Contact was obtained. The distance between the phenolic hydroxyl group of the substrate and the carbon atom on the sulfur of the SAM on the protein was also measured, and a reasonable conformation was selected to present the results given the reaction principle.
Single-site-directed mutagenesis and activity assay of 4VPMTs variants
4VPMT1 variants (S277A, I200A, M203A, V278A and Y61F) and 4VPMT2 variants (L138A, I164A, Y25F and H137A, F192A) were engineered by site-directed mutagenesis (QuikChange, Stratagene) using the primers in Supplementary Table 6. 4VPMT1 variants (F228A, H173A and W174A) and 4VPMT2 variants (L138W, Y173A, F238A and V242A) were engineered by SynbioB company. Expression and purification of all these variants were done according to the same protocol described for the wild type.
The activity assay of 4VPMT1 variants was carried out by pipetting 399 μl of the reaction mixture into a sample vial (RY-10100, 5 ml for headspace adsorption) containing buffer (20 mM Na2HPO4-NaH2PO4, 50 mM NaCl, pH 7.5), 20 μg 4VPMT1 variants and wild-type protein, and 4 μl SAM (100 mM); the reaction was started by adding 1 μl 4VP (0.4 mM), and the vial was then sealed with a cap (RY-10100, Sample bottle with clamp cap). The products were collected using SPME for 10 min at 30 °C and analysed by GC–MS. To determine the activity assay of 4VPMT2 variants, 399 μl of the reaction mixture was pipetted into a sample vial (RY-10100, 5 ml for headspace adsorption) containing buffer (20 mM Na2HPO4-NaH2PO4, 50 mM NaCl, pH 7.5), 180 μg 4VPMT2 variants and wild-type protein, and 4 μl SAM (100 mM); the reaction was started by adding 1 μl 4VP (30 mM), and the vial was then sealed with a cap (RY-10100, Sample bottle with clamp cap). The products were collected using SPME for 20 min at 30 °C. The amounts of product in the reactions were calculated according to the standard curve of 4VA.
Injection of 4NP
Two microlitres of 4NP aqueous solution with different concentrations (0.005 mM, 0.05 mM, 0.5 mM, 5 mM and 50 mM) were injected into the second abdominal segment of gregarious locusts. The effects of 4NP on 4VA production in locusts were investigated by GC–MS at 2 h after injection. Furthermore, 2 μl 4NP (0.5 mM) was injected into gregarious locusts, and 4VA was detected by GC–MS at 2 h, 4 h, 8 h, and 12 h. All control groups were injected with 2 μl H2O.
Feeding of 4NP with plants
4NP was dissolved in sterile water at 2 mM concentrations. One millilitre 4NP solution was sprayed uniformly on the stems and leaves of wheat seedlings. The fifth-instar nymphs were fed with wheat seedlings supplemented with 4NP. The control group was fed with wheat seedlings sprayed with sterile water. The volatiles of migratory locusts were collected and 4VA were quantified by GC–MS/MS after 24 h, 48 h and 72 h of feeding, respectively. The behavioural states of fifth-instar nymphs after 72 h of feeding were tested in a well-established behavioural assay arena.
Behavioural assay
The dual-choice behaviour assay was performed in a vertical airflow olfactometer as described1,24. To test the responses of locusts to the identified compounds, the diluted chemical was coated on filter paper (3 cm × 3 cm; Whatman No. 1), and mineral oil was used as a control agent in another funnel. To test the responses of locusts to the volatiles of 4NP-fed locusts, 30 4NP-fed gregarious locusts and 30 H2O-fed gregarious locusts were used as the odour sources of two sides of the chamber, respectively. A video camera (Panasonic), combined with VCR software (v.2, Noldus Information Technology), recorded locust behavioural activities within 10 min. The video was analysed using EthoVision XT software (v.11.5, Noldus Information Technology) to measure the total time spent on each side.
The phase behaviour assay was performed as previously described13,16. The experiment was performed in a rectangular perspex cage (40 cm length × 30 cm width × 10 cm height), where the top was clear and the walls were opaque. One of the separated chambers (7.5 cm length × 30 cm width × 10 cm height) contained 30 migratory locusts as the stimulation group, and the other was empty. The middle of the cage was the detection area. The locusts were released into the detection arena through a tunnel connected to the steel pipe at the centre of the detection area. The EthoVision video tracking system (Noldus Information Technology) automatically records individual behaviour. Each migratory locust was monitored for 6 min and tested only once. Five different behavioural parameters were extracted from the video: TDM, TDMV, total duration of stimulation area, total duration of relative area, and AI (AI = total duration of stimulation area-total duration of relative area). A binary logistic regression model described in the previous study was used to assess the phase status of the migratory locust. The regression model is as follows: Pgreg = eη/(1 + eη), where η = 2.11 + 0.005 AI + 0.012 TDM + 0.015 TDMV. Among them, Pgreg indicates the probability of a locust in the gregarious phase (Pgreg = 1 means the locust is fully gregarious, whereas Pgreg = 0 means individuals display solitary behaviour).
Single-sensillum recordings
To investigate the effects of 4NP on locust perception for 4VA, we conducted single-sensillum recordings of fifth-instar nymphs. Chemical substances such as single-sensillum recording stimulants included mineral oil as the blank, which was used to dilute 4VA by 1/10 (v/v). A piece of filter paper (Whatman) was placed in a 15 cm Pasteur glass tube and 10 μl of volatile solution was added to the filter paper. The responses of basiconic sensilla in 4NP-fed and control locusts were recorded and analysed as previously described1.
Computation-aided discovery of tolcapone as a 4VPMTs inhibitor
We conducted virtual screening on a library of 3,807 active small molecules using the GNINA software25 to identify potential target compounds. During the screening process, we first ranked all the small molecules based on their scores from low to high. To enhance the diversity of the screening results, we analysed the structural similarity of the top-scoring small molecules and excluded compounds sharing more than 0.55 similarity with previously selected compounds based on the ECFP fingerprint of the molecules calculated by RDkit. This ensured the selected compounds exhibited chemical diversity. Ultimately, 80 candidate molecules were chosen for the next round of screening.
In the second round of screening, we used CREST26 to determine the optimal conformation of each molecule and to compare these optimal conformations with their bound conformations. This step aimed to evaluate the conformational stability of the candidate molecules’ binding model and, in particular, exclude compounds with excessive torsional strain that might render them unstable. After rigorous screening, seven small molecules emerged as the most promising candidates and were selected for enzymatic activity assays following the same procedure described above.
Statistics and reproducibility
4VA levels in starvation treatment experiments were analysed using one-way ANOVA with Tukey’s multiple comparisons tests. 4VA levels in lignin feeding experiments were analysed using a two-tailed unpaired t-test. The differences in 4VA-d4, Phe-d5, CA-d6, p-HCA-d4 and 4VP-d4 between treatment and control groups were compared using a two-tailed unpaired t-test. The differential expressions of genes between gregarious and solitary locusts and 4VA levels in gene RNAi knockdown experiments were compared using a two-tailed unpaired t-test. The expression of 4VPMT1 and 4VPMT2 during the crowding and isolation process were analysed using one-way ANOVA with Tukey’s multiple comparisons tests. The enzymatic activities of 4VPMT1 and 4VPMT2 between mutants and wild type were compared using a two-tailed unpaired t-test. The inhibition effects of 4VP analogues on 4VPMT1 enzymatic activity were analysed using a two-tailed unpaired t-test. The emissions of 4VA in gregarious locusts injected with 4-nitrophenol and H2O were compared using a two-tailed unpaired t-test. The behavioural data of locusts in dual-choice assay were analysed by Wilcoxon signed-rank test (two-sided). The phase states of locusts (Pgreg) were analysed by the Mann-Whitney U-test (two-sided). Differences were considered significant at P < 0.05. Randomization and blinding details are provided in the Reporting Summary. Each experiment was repeated three times independently with similar results. All statistics were analysed using SPSS 18.0 (SPSS Inc., Chicago, IL, USA). All the statistical results throughout this study are listed in Supplementary Table 7.
Ethics declaration
Animal experiments were approved by the Animal Care and Use Committee of the Institute of Zoology, Chinese Academy of Sciences, or Peking University.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.