Generation of scaffold library of cyclic Y-state oligomers
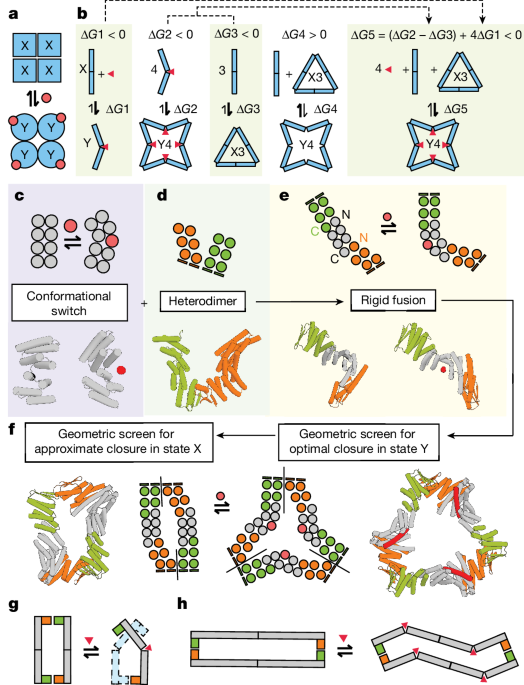
Cyclic oligomers were constructed through the modular fusion of two classes of de novo proteins: (1) three hinge proteins (cs074, cs221, js007) that were previously confirmed to switch between two defined conformational states, âXâ and âYâ, in response to peptide binding11 and (2) heterodimeric alpha-beta proteins (LHDs) that were designed to reversibly associate and dissociate in dynamic equilibrium12. To create helical surfaces that would facilitate modular fusion between LHDs and hinges, and to sample a diversity of possible angular fusions between these components, we used the HFuse26 software to computationally generate a library of designed helical repeat (DHR) fusions to each monomer within an LHD (an example is shown in Extended Data Fig. 1b). The junctions between LHDs and DHRs were then redesigned with Rosetta FastDesign with backbone movement54 to stably pack the junctions between them. These designs were then predicted with alphafold v.2 (ref. 28) (AF2), and we filtered for designs that reported a model pLDDT (predicted local distance difference test) score greater than 88 and C-α r.m.s.d. (root mean square deviation) less than 2.5âà to the original HFuse backbone. Dimer LHDâDHR fusions with two helical termini available for further fusion, one at the C terminus and the other at the N terminus, were then used as building blocks in the subsequent steps, with each terminus being used as a surface for fusion to the N and C termini of a hinge (Fig. 1c). The WORMS26 software was used to generate a library of rigid fusions between the LHDs and hinges (in their peptide-bound âYâ state) that would result in cyclic closure into a C2, C3, C4 or C5 oligomer in the presence of peptide. Splicing of up to two terminal helices on the hinges and 150 terminal residues on the LHDâDHR monomers was permitted during the WORMS-fusion process to generate robust junctions, while preserving binding function at both LHD and hinge interfaces. âYnâ oligomers with a monomer length of fewer than 450 amino acids were selected for further design. For each peptide-bound Y-state monomer generated by WORMs, we generated models of the X-state structure that it would adopt in the absence of peptide. We did this by aligning segments of the Y-state chimeric protein, N and C-terminal to the hinge region, to the known X-state of the corresponding parent hinge. This yielded a batch of X-state models that could be used for comparisons in AF2 filtering in subsequent steps.
ProteinMPNN redesign of junctions and AF2 filtering for fusion quality
To ensure the solubility and rigidity of these fusions, we redesigned the residues at the WORMS-generated junctions with ProteinMPNN27, while fixing the amino-acid identities of residues (1) key to the conformational switching of the hinge and (2) that mediate assembly into rings at the LHD interfaces. The former category included residues that directly interact with the peptide at the cleft as well as ones that assist in packing the backside of the hinge when the peptide is bound (Extended Data Fig. 1a). Four sequences were generated per design using ProteinMPNN. These sequences were then predicted as monomers with AF2 in the absence of the peptide with three recycles, yielding putative X-state structures. We filtered for ProteinMPNN-designed ring proteins with pLDDT greater than 86, and a C-α r.m.s.d. of under 2.75âà to the X-state design model that was generated as described above.
Prediction of X-state oligomers
To generate the predicted X-state oligomer (Xm), for each design that passed this initial filter, we iteratively docked five AF2-predicted monomers end-to-end along their LHD interfaces to generate a series of oligomers, from dimer to pentamer. We recorded the distance between the N-terminal LHD domain of the first monomer and the C-terminal LHD domain of the nth monomer in the chain at each docking step by computing the shortest distance between all possible pairings of backbone carbon atoms across the two domains (Extended Data Fig. 2a). Oligomeric states in which this atomic distance is minimal were identified to measure the closest approach of the X-state oligomer to cyclic closure. We filtered for designs in which this distance was less than 24âà to enrich for designs that are likely to close in the X-state rather than extend into filaments (Extended Data Fig. 2b). A subset of these filtered designs was then manually selected for experimental characterization to span a range of symmetries, shapes and cognate effectors. To determine the extent to which the monomers must individually bend to mediate closure in the X state, we predicted them in their assembled form using alphafold multimer v.3. We used rigid docks of the X-state AF2-predicted monomers prepared in the previous step as templates for prediction with three recycles. In 23 out of 26 cases, the monomers underwent some degree of flexible deviation away from the X-state to close the ring with optimally or near-optimally satisfied LHD interfaces. To quantify this deviation in the closed cases, we measured the r.m.s.d. between the ring-incorporated monomer and the free monomer predictions, as shown in Extended Data Fig. 2c. AF2 multimer v.3 did not predict C4 oligomers or higher as closing into rings, preferring to model them as dihedrals with unsatisfied LHD interfaces.
Design of static control rings
To assess the importance of designing junctions to target optimal closure in the Y state, but suboptimal closure in the X state, we implemented a modified design procedure in which the unbound X-state ring is optimized for closure, rather than the Y-state ring. We used WORMS to screen for LHD-hinge fusions that resulted in the perfect end-to-end closure of an X-state cyclic ring containing between three and five subunits. ProteinMPNN was used to design the junctions between the components while preserving the key functional residues in the hinge domain, as described above. AF2 was used to predict the monomer structures and validate the fusion junction designs. Designs with a pLDDT greater than 86 and AF2-predicted C-α r.m.s.d.â<â1.5âà to X-state design were selected for further characterization.
Design of double-hinge rings that retain symmetry on peptide binding
To design rings that can toggle between different oligomeric conformations without changing their original oligomeric state, we first docked two cs221 hinges together such that the C-terminal helices of the first hinge contact the N-terminal helices of the second hinge (Extended Data Fig. 7b). This was done by manual placement of these two domains end-to-end in PyMOL58 along various angles of approach, such that the peptide binds to opposite sides of the fusion construct. We used RFDiffusion33 to build a short loop region connecting both hinges into a single chain. ProteinMPNN27 was used to redesign the interface between these two hinge domains, without changing the sequence of the rest of the protein. We then predicted these fusions using AF2 and filtered for designs that predicted with a pLDDTgreater than 0.90 and C-α r.m.s.d.â<â1âà . These were then used as inputs into the same WORMs-based fusion strategy described above, specifically targeting closure into C2 rings in the Y state (Extended Data Fig. 3aâc). We used AF2 to predict the resultant LHD-hingeâhinge-LHD outputs in the X-state, isolating designs with a pLDDT greater than 0.85 and C-α r.m.s.d.â<â1.5âà . We then docked the X-state monomers to confirm that the filtered designs approach C2 symmetry in the absence of peptide. We manually selected a set of 24 for further characterization.
Design of inducible homodimers
The WORMS-based fusion approach described above was used to target a C2-symmetric homodimeric assembly in state Y, through the fusion of LHDâDHRs to hinges. ProteinMPNN was used to redesign the junctions of the monomeric subunits, followed by folding with AF2 in the absence of peptide to generate an X-state monomer. We filtered for designs that were predicted with a C-α r.m.s.d.â<â2.75âà to the expected X-state backbone and pLDDT greater than 86. After this, the X-state predicted monomers were docked along one of their LHD interfaces. We used Rosetta55 to filter for designs with backbone clashes (Rosetta fa_rep score greater than 10,000) in this docked state, corresponding to a sterically prohibited dimer form. The filtered designs were manually inspected to ensure that the LHD produced substantial backbone clashes in the X state, and a subset of 12 were then experimentally characterized.
Design of symmetric C3 and C5 subcomponents for dihedral assemblies
The WORMS protocol was used, without symmetry constraints, to generate rigid fusions of hinges to previously validated C3 (ref. 44) and C5 (ref. 45) oligomers. The C terminus of hinge cs221 was rigidly fused to these cyclic oligomers in an orientation that ensured the N terminus of the hinge would be accessible for dihedral docking in state X. Fusion junctions between hinges and cyclic oligomers were then redesigned with ProteinMPNN while preserving the oligomeric interfaces of the parent scaffolds along with key functional residues of the hinge protein. To increase the number of oligomers for downstream docking applications, eight sequences were generated for each rigidly fused oligomer. Redesigned proteins were then predicted as monomers using AF2 with initial guess56 and designs with a pLDDT greater than 85 and an r.m.s.d.â<â2.5âà were selected for dihedral docking.
Docking of subcomponents
RPXDock46 was used to sample D3 and D5 assemblies containing two copies of a C3 or C5 hinge fusion, respectively. Docking was guided towards the first 36 residues of component monomers, such that the N-terminal helices of the opposing cyclic oligomers packed against one another along a dihedral plane of symmetry. Docked designs were then sequence optimized along the dihedral interface using ProteinMPNN with the tied residues feature, such that the interface for each chain pair along the dihedral axis contained identical residues. To evaluate and filter for these newly designed dihedral interfaces we extracted C2-symmetric dimeric subunits and predicted their structure with AF2 with an initial guess56. Designs passing metrics of pLDDT greater than 85 and C-α r.m.s.d.â<â2.5âà were then chosen for further characterization.
Recombinant expression and purification
Genes were codon-optimized for expression in Escherichia coli (E. coli). DNA fragments encoding designed proteins were ordered as eblocks from IDT and cloned into custom plasmids bearing a T7-promoter driven expression system with a C-terminal sequence-specific nickel assisted cleavage site (SNAC tag)57 and 6xHis-tag (âProtein-GSHHWGSTHHHHHHâ) using Golden Gate Assembly. All proteins were expressed in NEB BL21(DE3) E. coli cells using TBII (MpBio) autoinduction media, which was supplemented with ZYM-5052, trace metal mix, 2âmM MgSO4 and 50âmgâmlâ1 Kanamycin. 50âml of expression cultures were grown at 37â°C for 6âh followed by 20â°C for 24âh with shaking at 225ârpm throughout.
Cells were then collected by centrifugation at 5,000g and resuspended in 15âml of Tris-buffered saline (TBS) lysis buffer (300âmM NaCl, 40âmM Tris, 40âmM Imidazole, pH 8). Cells were lysed by sonication in the presence of 1âmM DNase, 1âmM Pierce Protease Inhibitor Mini Tablets, EDTA-free per 100âml and 1âmM PMSF added immediately before lysis. Cell debris was pelleted by centrifugation at 20,000g for 40âmin. The supernatant was then added to roughly 1âml of Ni-NTA Metal affinity chromatography resin to separate the protein from impurities in a vacuum manifold. The protein was washed with 10à bead volume of TBS (300âmM NaCl, 40âmM Tris, 40âmM imidazole pHâ8) and protein was eluted in 2âml of 300âmM NaCl, 40âmM Tris, 500âmM imidazole. Eluted protein was then further purified with SEC on an automated fast protein liquid chromatography (FPLC) system using Superdex 200 Increase 10/300 GL columns in TBS (40âmM Tris, 300âmM NaCl, pHâ8) with 1âml fractions. Final concentrations were estimated using ultraviolet absorbance at 280ânm (UV280) with a NanoDrop 2000/2000c, relying on molar extinction coefficients and molecular weights predicted from the sequence. To confirm the protein sequence, we measured the molecular mass of each protein by mass spectrometry; intact mass spectra were obtained by reverse-phase liquid chromatograhy with mass spectrometry on an Agilent G6230B TOF on an AdvanceBio RP-Desalting column, and subsequently deconvoluted by way of Bioconfirm using a total entropy algorithm. Sequences picked for further characterization beyond SEC-binding assays were re-ordered as precloned genes from IDT in pet29B expression vectors. The cognate peptides for these proteins (cs074B, cs221B and js007B) were chemically synthesized as previously described11.
In constructs with designed disulfides (sr312_y_staple), the expression and purification were the same as described above, with two modifications: (1) 1âmM TCEP (tris(2-carboxyethyl)phosphine) was added to the lysis buffer to prevent premature disulfide formation during purification, (2) copper phenanthroline was added to the immobilized metal ion affinity chromatography elution at a final concentration of 10âmM and the resulting mixture was incubated overnight to encourage full formation of the disulfides.
SEC-binding experiments
To determine the effects of peptide binding on oligomerization state, we measured shifts in SEC profile in the presence and absence of roughly 2à molar excess of peptide (Fig. 2 and Extended Data Fig. 4). Protein assemblies and peptide were diluted into TBS (300âmM NaCl, 40âmM Tris, pHâ8) to a final monomer concentration of 5âμM and a final peptide concentration of 10âμM in a final volume of 700âμl. Peptide-free and peptide-bound samples were injected serially using an automated FPLC system (AKTA Pure) with a flow rate of 0.5âmlâminâ1 on a Superdex 200 Increase 10/300 GL column. Absorbance signals at UV230 and UV280 were measured to monitor the elution profile of protein across the run. For constructs that included a GFP tag, UV473 absorbance was also measured. For the SEC titration series of sr312 shown in Extended Data Fig. 7a, the unbound fraction was calculated by estimating the area under the X3 peak in each measurement with UNICORN v.7.3 and dividing it by the area under the X3 peak in the absence of the effector peptide (in which 100% of the protein is unbound). The bound fraction was then calculated by subtracting this value from 1. SEC traces of several runs that are shown as overlays were run subsequently on the identical FPLC system and column, on the same day and using the identical buffer to ensure ideal comparability.
Fluorescence polarization
All fluorescence polarization binding assays were conducted in 96-well black-bottom microplates (Corning, catalogue no. 3686) at room temperature (roughly 25â°C). Fluorescence measurements, including parallel intensity, perpendicular intensity and polarization, were taken in a Synergy NEO2 plate reader, with a 530/590 filter cube. For each design, four replicate titration series were prepared per plate by serial twofold dilution of a starting stock of 20âμM protein into TBS (300 NaCl, 40âmM Tris, pHâ8.0 buffer) across 24 wells. TAMRA-labelled peptide was kept across this series at a constant concentration of 1ânM. The final volume of peptide-protein mixture in each well was kept constant at 80âμl. The measured polarization signal was fitted to the following equation to determine Kd.
$${\rm{F}}{\rm{P}}\,{\rm{s}}{\rm{i}}{\rm{g}}{\rm{n}}{\rm{a}}{\rm{l}}=\frac{{\rm{m}}{\rm{a}}{\rm{x}}{\rm{i}}{\rm{m}}{\rm{u}}{\rm{m}}\,{\rm{s}}{\rm{i}}{\rm{g}}{\rm{n}}{\rm{a}}{\rm{l}}({V}_{{\rm{m}}})\times {[{\rm{r}}{\rm{i}}{\rm{n}}{\rm{g}}{\rm{m}}{\rm{o}}{\rm{n}}{\rm{o}}{\rm{m}}{\rm{e}}{\rm{r}}{\rm{c}}{\rm{o}}{\rm{n}}{\rm{c}}{\rm{e}}{\rm{n}}{\rm{t}}{\rm{r}}{\rm{a}}{\rm{t}}{\rm{i}}{\rm{o}}{\rm{n}})}^{n}}{{{K}_{{\rm{d}}}}^{n}+{[{\rm{r}}{\rm{i}}{\rm{n}}{\rm{g}}{\rm{m}}{\rm{o}}{\rm{n}}{\rm{o}}{\rm{m}}{\rm{e}}{\rm{r}}{\rm{c}}{\rm{o}}{\rm{n}}{\rm{c}}{\rm{e}}{\rm{n}}{\rm{t}}{\rm{r}}{\rm{a}}{\rm{t}}{\rm{i}}{\rm{o}}{\rm{n}}]}^{n}}+{\rm{b}}{\rm{a}}{\rm{s}}{\rm{e}}{\rm{l}}{\rm{i}}{\rm{n}}{\rm{e}}$$
where b, Vm, n and Kd are fit by nonlinear regression to a set of polarization signal and protein concentration values using the optimize_curvefit function within SciPy. An average of Kd from four replicates and their standard error are reported in Extended Data Fig. 6.
TAMRA SEC on sr312
TAMRA-labelled peptide at varying concentrations (10, 5, 2, 1, 0.5, 0.25âμM) was separately mixed with a constant 1âμM concentration of sr312 in a final volume of 200âμl in TBS (40âmM Tris, 300âmM, pHâ8.0). Mixtures were serially injected with an autosampler on a high-performance liquid chromatography system (Agilent 1260 Infinity II LC). In addition to absorbance at UV280, TAMRA fluorescence at 590ânm was recorded with an excitation wavelength of 570ânm to specifically monitor the elution of labelled peptide. SEC traces were collected over a 9âmin interval from a Superdex 200 GL5/150 Increase column at a flow rate of 0.35âmlâminâ1 in TBS (300 NaCl, 40âmM Tris, pHâ8). Finally, 100âμl fractions were collected across the elution run time.
Characterization of complexes by MP
All MP measurements were carried out in a TwoMP (Refeyn) Mass photometer. For initial characterization of rings in Fig. 2, protein and peptide were incubated at 1âμM protein concentration, with or without 10âμM peptide, for 20â25âh at room temperature to allow the system to reach equilibrium. For MP, data are shown in Fig. 5. Dihedral samples were incubated at 5âμM with 2à molar excess of either cs221B or effector protein 3hb21 overnight. Samples were then diluted to 50â200ânM monomer concentration (roughly 20ânM oligomer) immediately before measurement to limit overcrowding of the field of view. A 12-well gasket was placed on each slide. Then 10âμl of buffer was added to one well of this gasket and the camera was brought into focus after orienting the laser to the centre of the sample well. Next, 10âμl of sample was added to this droplet and 1âmin videos were collected with either a large field of view (for ring and dihedral complexes) or a small field of view (for inducible homodimers) in AcquireMP. Ratiometric contrast values for individual particles were measured and processed into mass distributions with DiscoverMP. For each design, a sample of 20ânM Beta-amylaseâconsisting of monomers (56âkDa), dimers (112âkDa) and tetramers (224âkDa) in equilibriumâwas used to arrive at a mass calibration; thereby allowing contrast values to be converted into mass values across tested designs. Expected masses for Xm and Yn species were calculated by multiplying liquid chromatograhy with mass spectrometry estimated monomer masses by the number of subunits in different oligomeric configurations. Distributions were exported from DiscoverMP and plotted with a custom script in Python. Gaussian distributions were fit to this peak to estimate observed oligomer masses and mass error using normfit in the SciPy package (Extended Data Fig. 5).
For sr312 cooperativity measurements in Fig. 4, a titration series of GFP-tagged peptide (cs221B) spanning 10 to 0.5âμM was mixed with a constant concentration of 2âμM sr312. For sr508 cooperativity measurements, GFP-tagged peptide (cs221B) spanning 2 to 0.1âμM was mixed with a constant concentration of 250 nM sr508. Solutions were incubated and measured as described. DiscoverMP was used to fit Gaussian distributions to bound and unbound species in the mass distributions, with three technical replicates for each mixture. Proportions of bound and unbound hinge could then be estimated with the following equation:
$${\rm{B}}{\rm{o}}{\rm{u}}{\rm{n}}{\rm{d}}\,{\rm{f}}{\rm{r}}{\rm{a}}{\rm{c}}{\rm{t}}{\rm{i}}{\rm{o}}{\rm{n}}=\frac{4\times [Y4P4\,{\rm{c}}{\rm{o}}{\rm{u}}{\rm{n}}{\rm{t}}{\rm{s}}]+2\times [Y4P2\,{\rm{c}}{\rm{o}}{\rm{u}}{\rm{n}}{\rm{t}}{\rm{s}}]}{4\times [Y4P4\,{\rm{c}}{\rm{o}}{\rm{u}}{\rm{n}}{\rm{t}}{\rm{s}}]+4\times [Y4P2\,{\rm{c}}{\rm{o}}{\rm{u}}{\rm{n}}{\rm{t}}{\rm{s}}]+3\times [X3\,{\rm{c}}{\rm{o}}{\rm{u}}{\rm{n}}{\rm{t}}{\rm{s}}]}$$
The SciPy optimize_curvefit function was used to fit an occupancy curve to the data collected, allowing for estimation of apparent Kd and Hill coefficient (n) using the Hill equation, where θ refers to the fraction of protein bound to ligand:
$$\theta =\frac{{[L]}^{n}}{{K}_{{\rm{d}}}+{[L]}^{n}}$$
nsEM on switchable rings and dihedral complexes
Carbon-coated 400 mesh copper grids (01844-F, TedPella, Inc.) were glow discharged using a PELCO easiGlow Glow Discharged Cleaning System. SEC-purified proteins were diluted to 0.01âmgâmlâ1 with SEC buffer (300âmM NaCl, 40âmM Tris, pHâ8.0), and then immediately pipetted onto the glow-discharged grids. The protein solution was allowed to sit on the grid for 1âmin, before being blotted away with Whatman filter paper. Then 3âμl of 2% uranyl formate stain was added to the grid and then blotted away after 1âmin. A second and third wash of uranyl formate stain were added to the grid, allowed to sit for 30âs each, before being blotted away. The grid was allowed to air-dry for 5âmin. Dried grids were then imaged using a FEI Talos L120C TEM (FEI Thermo Scientific) equipped with a 4,000âÃâ4,000 Gatan OneView camera, at a magnification of Ã57,000 and pixel size of 2.49âà . Once a grid-square with satisfactory stain thickness and contrast was identified, EPU software was used to automatically collect 200â400 micrographs across the square. Micrographs were imported into and analysed using CryoSPARC v.4.0.3. Patch CTF was used to estimate defocus variation for micrographs. Given the radius of particles, CryoSPARC automatically picked and extracted particles from the contrast transfer function (CTF) corrected micrographs. Particles then were subjected to 2D classification to find 2D averages that could be used as templates for more precise particle picking across the CTF corrected micrographs. After particle picking and extraction from micrographs, a further round of 2D classification was done to find higher resolution averages of the oligomers in various states and orientations.
FRET
FRET constructs for IHA10 were designed by picking two sites on opposing sides of the dimer interface and separately mutating each to cysteine in two point-mutant constructs, IHA10_8C and IHA10_323C. FRET labelling sites were chosen to be close enough to maximize signal in the dimer form (Extended Data Fig. 9g), while being far enough from the interface to not sterically block assembly when conjugated with the fluorescent dye. DNA constructs encoding these two point mutants were ordered from IDT as precloned genes in the same expression vector described above. As donor and acceptor dyes, we used AlexaFluor 555 C2 maleimide (donor) and AlexaFluor 647 C2 maleimide (acceptor), which were purchased from Thermo Fisher Scientific. Next, 1âmg of each dye was dissolved in 200âμl of DMSO to yield a stock solution at 5âmM. Cysteine mutants were expressed and purified according to the previously described procedure, with a modification that 0.5âmM TCEP was added to the buffer during lysis, immobilized metal ion affinity chromatography and SEC. Furthermore, 20âmM sodium phosphate (pHâ7.0) instead of Tris-HCl was used as a buffer during SEC. Following the SEC step, a 500âμl solution each of IHA10_8C and IHA10_323C at a concentration of 50âμM was subjected to 2âh of incubation at room temperature with 500âμM of a single dye (AlexaFluor 555 and AlexaFluor 647, respectively). The labelled samples were then purified by SEC to eliminate excess dye, in a buffer of 20âmM Tris-HCl and 300âmM NaCl at a pH of 8. The FRET titration was conducted at 25â°C in 96-well plates (Corning 3686) using a Synergy Neo2 plate reader. The excitation wavelength was 520ânm and emission wavelength was 665ânm (Fig. 5f). A response curve was fitted to the data by nonlinear regression with a custom python script.
Luciferase assay for inducible homodimers
Gene fragments encoding inducible dimer designs were ordered as eblocks from IDT and cloned into custom plasmids that included either a C-terminal fusion to the lgBit subunit of NanoLuc or an N-terminal fusion to the smBit of NanoLuc. Ordered sequences also included a C-terminal Histidine tag. As a control for the js007-based hinge designs, we also tagged the corresponding parent hinge, js007A, with each of these subunits to ensure that the luciferase signal is driven primarily by assembly. Proteins were expressed and purified as described above. Luciferase assays were performed in 40âmM Tris-HCl, 300âmM NaCl, pHâ8, 0.05% v/v Tween 20. Reactions were assembled in 96-well plates (Corning 3686) and luminescence was measured in a Synergy Neo2 plate reader (BioTek). LgBit- and smBit-fused constructs were mixed at equimolar concentrations that ranged from 5 to 20ânM (reported in figure legend) in a final volume of 80âμl. Mixtures were incubated at room temperature overnight to ensure that the system reaches equilibrium. For single-peptide-concentration comparisons shown in Fig. 5e, effector peptide was added at a concentration of 20âμM and ten replicates were collected for this sample as well as all controls. For titrations shown in Extended Data Fig. 9b, a twofold dilution series was prepared from a starting concentration of 10âμM, and spanned 24 concentrations. The average of four technical replicates across one plate is shown. In all cases, 10âμl of Nano-Glo substrate at 10à dilution was added immediately before measurement, with a dead time of around 10â30âs.
Cryo-EM sample preparation
To prepare the samples, 2âμl of sr322 with js007 effector peptide (sr322_ js007B), sr312 with cs221B effector peptide (sr312_cs221B), sr322 at 0.971âmgâmlâ1 in 150âmM NaCl, 40âmM Tris, pHâ8, was applied to glow-discharged C-flat holey carbon grids. Vitrification was performed using a Mark IV Vitrobot at 4â°C for sr322_ js007B and sr312_cs221B, and 22â°C for sr322 with 100% humidity for all. Samples were frozen on glow-discharged 2.0/2.0-T C-flat holey carbon grids for sr322_ js007B and sr312_cs221B, 1.2/1.3-T C-flat holey carbon grids for sr322. Blotting was done using a 5.5âs blot time, a blot force of 0 and a 5âs wait time for sr312_cs221B and sr322_ js007B; a 6.5âs blot time, a blot force of 0 and a 7.5âs wait time for sr322 was used before being immediately plunge frozen into liquid ethane.
Cryo-EM data collection
sr322_ js007B, sr322 and sr312_cs221B were collected automatically using SerialEM58 and used to control a ThermoFisher Titan Krios 300âkV TEM for sr322 and sr312_cs221B and a ThermoFisher Glasios 200âkV TEM. Both microscopes were equipped with a standalone K3 Summit direct electron detector59 and operated in super-resolution mode for sr312_cs221B and counting mode for sr322_ js007B and sr322. Random defocus ranges spanned between â0.8 and â1.8âμm using image shift, with one shot per hole and nine holes per stage move. Altogether, 1,398, 3,795 and 4,213 videos with pixel sizes of 0.885, 0.4215 and 0.843 and doses of 50, 43 and 52âeâ/à 2 were recorded, respectively, for sr322_ js007B, sr312_cs221B and sr322.
Cryo-EM data processing
All data processing was carried out in CryoSPARC60. The video frames were aligned using Patch Motion with an estimated B factor of 500âà 2. The maximum alignment resolution was set to 3. Outputs were binned to a final pixel size of 1.0288âà per pixel by setting the output F-crop factor to one half. Defocus and astigmatism values were estimated using the Patch CTF with the default parameters. In total, 1,614,340 particles were picked in a reference-free manner using Blob Picker and extracted with a box size of 340 for sr322; for sr312_cs221B and sr322_ js007B, a manual picker was first used to pick 590 particles and 2,804 particles with box sizes of 400 and 256, respectively. An initial round of reference-free 2D classification was performed in CryoSPARC using 150, 50 and 50 classes and a maximum alignment resolution of 6âà for sr322, sr312_cs221B and sr322_ js007B, respectively. The best classes were next low-pass filtered to 20âà and used as templates for a second round of particle picking using Template Picker, resulting in a new set of 996,592, 971,294 and 524,968 particle picks that were extracted with box sizes of 340, 600 and 300âpixels for sr322, sr312_cs221B and sr322_ js007B, respectively. For sr322_b11, only top views along C5-symmetric access were seen, The best 2D class averages were shown with 66,770 particles. For sr322 and sr312_cs221B, 996,592 and 971,294 particles were then used for 3D ab initio determination using the C1 symmetry operator. Initial ab initio showed density for a clustered species with sr322, and for sr312_221B a preferred orientation failed to produce a good map. To further resolve sr322, clustered species particles were reextracted with a 400âpixels box size, Fourier cropped to 200âpixels. Using 279,729 particles, a three-class ab initio was run in C1 to sort monomeric and clustered species. For the final refinement of the sr322 clustered species, 144,551 particles were further processed using non-uniform refinement in C2 with a final estimated global resolution of 4.32âà . Another round of template picking was used to pick out monomeric sr322 and to pick out more side views for sr312_cs221B with 924,961 and 1,485,952 particles with box sizes of 340 and 600, respectively, were next funnelled into another round of reference-free 2D classification for sr322, with the best 157,386 particles submitted for homogenous refinement in the presence of C1 symmetry. The estimated global resolution of this map was determined to be 6.54âà . Once symmetry was confirmed in C1, these maps were refined further using homogenous refinement in C4 symmetry to an estimated global resolution of 4.55âà . For sr312_221B, several rounds of 2D classification were performed to remove excess top views and better classify side views with a final total particle count of 58,251. At this point, an ab initio was generated in C1 in high agreement with the design model and revealing excellent orientational sampling of the input particles. This map was refined with non-uniform refinement and achieved a final estimated global resolution of 4.40âà . These maps were refined with DeepEMhancer61 for sr322, the sr322 clustered species and sr312_cs221B. Local resolution estimates were determined in CryoSPARC using a Fourier shell correlation threshold of 0.143. 3D maps for the two half-maps, the final unsharpened maps and the final sharpened maps were deposited in the Electron Microscopy Data Bank (EMDB) under accession numbers EMD-42442, EMD-42491 and EMD-42542.
Cryo-EM model building and validation
The design model of sr322 and sr312_cs221B was used as an initial reference for building the final cryo-EM structures. PyMOL53 and UCSF Chimera62 were initially used to break apart the monomeric components and fit them in density. We then further refined the structure using the molecular dynamics flexible fitting simulation Namdinator63. This process was repeated iteratively until convergence and high agreement with the map was achieved. Several rounds of relaxation and minimization were performed on the complete structures, which were manually inspected for errors each time using Isolde64 and Coot65,66. Phenix67 real-space refinement was subsequently performed as a final step before the final model quality was analysed using MolProbity68. Figures were generated using UCSF ChimeraX62. The final structures were deposited in the Protein Data Bank (PDB) under 8UP1, 8URE and 8UTM.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.