Single-cell data collection and preprocessing of healthy samples
To assemble a comprehensive pan-tissue cell atlas, we collected scRNA-seq datasets and conducted quality control procedures via the Scanpy48 toolkit, as detailed in subsequent sections (Extended Data Fig. 1 and Supplementary Table 1). Default parameters were used unless otherwise specified.
Data collection
We included scRNA-seq datasets from adult samples that met the following criteria: (1) utilization of fresh, not frozen, samples; (2) inclusion of samples based on cell-type enrichment: (a) no cell-type enrichment; (b) a mixture of immune, epithelial, endothelial and stromal compartments; (c) enrichment for either immune or non-immune cell populations; and (3) generation of single-cell, not single-nucleus, data using the 10x Genomics platform. These criteria were implemented to minimize batch effects across the datasets49. Ultimately, a total of 33 datasets from 26 cohorts were included, collectively representing a cell atlas across 35 human tissues.
Quality control
To standardize datasets annotated with different versions of the human genome assembly, we limited the transcriptome to the common set of 21,812 genes found in the three most widely used 10x Genomics gene annotations, specifically GRCh38 (Ensembl 84), GRCh38 (Ensembl 93) and GRCh38 (GENCODE v32/Ensembl 98). Cells identified as low-quality or germ line in the original studies were excluded, and only cells meeting the following criteria were retained: 500–8,000 genes, 1,000–100,000 gene counts, and less than 20% mitochondrial gene counts. We applied Scrublet50, integrated into Scanpy, to each cohort and removed cells with a doublet score exceeding the 90th percentile across all cohorts. We then excluded samples with fewer than 50 high-quality cells. In the end, the analysis comprised a total of more than 700 samples that passed the stringent quality control measures.
Preprocessing
Beginning with the combined gene count matrix across all datasets, we derived the normalized gene expression matrix by normalizing total counts per cell (library size) using a scale factor of 10,000 followed by logarithmic transformation. Highly variable genes (HVGs) were then selected using the function scanpy.pp.highly_variable_genes with the following parameters: (n_top_genes=2000, flavor = “cell_ranger”, batch_key = “datasetID”). Notably, HVG selection was performed after removing specific genes, including immunoglobulin genes, T cell receptor genes, ribosome protein-coding genes, heat shock proteins-associated genes, and mitochondrial genes. Several confounding effects, including total gene counts per cell, the percentage of mitochondrial gene counts, and cell cycle were addressed, using the function scanpy.pp.regress_out. Finally, HVGs were centred and scaled among all cells.
Single-cell data integration and annotation
To integrate these extensive datasets, we used the Scanpy toolkit with default parameters unless otherwise specified.
Benchmarking integration methods
To determine the best integration method for our datasets, we utilized scIB17 to benchmark several widely used Python-based tools: BBKNN18, Harmony51, Scanorama52 and deep learning-based scVI53, scANVI54, and SCALEX55. Among the 14 metrics in scIB, biological conservation for HVG and trajectory were not applicable, and the kBET metric was excluded owing to memory requirements exceeding 2 TB. Overall scores were calculated as a weighted mean (40/60) of batch correction and biological variance conservation. Importantly, we conducted two independent benchmarking analyses on the entire atlas and a subset atlas, respectively. In the end, BBKNN emerged as the top performer and was used for the integration of the pan-tissue datasets (Extended Data Fig. 2).
Dataset integration
Principal component analysis was performed on the centred and scaled HVG expression matrix to extract 50 principal components. BBKNN, integrated into Scanpy, was then executed with the dataset as the batch variable. The batch-corrected graph was then utilized to perform UMAP56 for visualizing cells on a two-dimensional layout.
Cell clustering and annotation
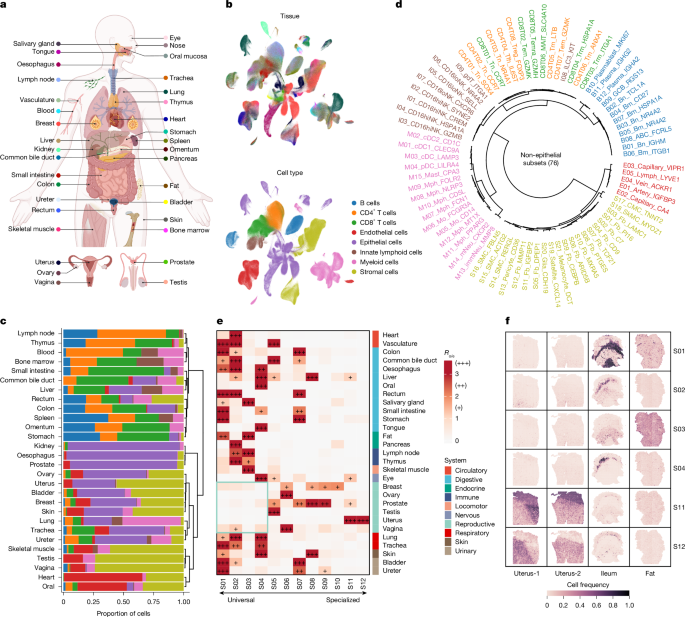
We performed at least two levels of unsupervised cell clustering and annotation. The first level of clustering was performed using the function scanpy.tl.leiden with resolution = 0.1 followed by identification of differentially expressed genes (DEGs; log2-transformed fold changes >1, FDR < 0.05, Student’s t-test). The eight broad cell types were identified on the basis of canonical markers and DEGs. We also received assistance with cell annotation from CellTypist7, an automated cell-type annotation tool, using the Immune_All_High and Immune_All_Low models. Subsequently, further clustering (second or more levels) was performed using context-specific resolutions to obtain several distinct cell subsets for each cell type. Epithelial cells were excluded from further clustering owing to their highly tissue-specific nature. In total, 2,293,951 high-quality cells from 706 samples across 317 donors were annotated into 76 non-epithelial subsets and 26 epithelia cell types.
Hierarchical clustering of cell subsets
We generated pseudo-bulk profiles for 76 non-epithelial cell subsets by averaging the gene expression of all cells within the same subsets. Next, unsupervised hierarchical clustering was performed using correlation distance and the hclust function (method = “ward.D”). The results were visualized using the dendextend R package.
CoVarNet framework
We introduced CoVarNet, a computational framework designed to systematically unravel coordination among multiple cell types. CoVarNet identifies co-occurring CM networks by analysing the covariance in cell subset frequencies across various samples.
CoVarNet overview
CoVarNet uses input data on cell subset frequencies within each cell type and sample. It utilizes two parallel modules to jointly determine CM networks by connecting co-occurring subsets (nodes) through edges. The first module applies NMF to the cell subset frequency matrix, identifying factors that prioritize subsets based on their weights. The top subsets of each factor act as co-occurring nodes in a single CM network. The second module identifies specifically correlated subset pairs, which act as potential edges. Multiple CM networks are then constructed to interconnect co-occurring nodes via these potential edges, followed by topological and statistical evaluations.
Input frequency matrix
To ensure comparability of cell-subset frequencies across tissues and clinical specimens, we included only samples without cell-type enrichment or those from mixtures of the four cell compartments. Samples with fewer than 50 high-quality cells were excluded. For each eligible sample, we computed the frequencies of cell subsets within their corresponding cell types. Min-max normalization was applied to correct the frequency matrix, mitigating the impact of varying numbers of cell subsets across different cell types. Thus, a corrected frequency matrix, ranging from 0 to 1, was utilized in the CoVarNet procedure. Specifically, we generated a frequency matrix consisting of 76 subsets (rows) and 510 samples (columns) for the pan-tissue atlas.
NMF
NMF has been used in the analysis of single-cell57,58,59,60 and spatial61,62 expression data to extract gene expression programmes. In this study, CoVarNet applies NMF to the frequency matrix to decipher cellular co-occurring programmes, specifically using the nsNMF method with ranks from 2 to 20, as implemented in the NMF R package63. To ensure robustness, we conducted 30 runs to derive a consensus output, consisting of k factors and their activities in each sample. Specifically, the top ten subsets of each factor were used as co-occurring node candidates in a single CM network for the pan-tissue analysis.
Rank selection
To determine the optimal rank for NMF analysis, we used the cophenetic correlation coefficient (CCC) as the evaluation index, in accordance with practices from previous reports1. CCC is used to quantify classification stability, with values ranging from 0 to 1 and 1 indicating maximum stability64. We denoted the CCC at rank k as ρk and established a procedure tailored to this context for consistent stability based on the following criteria: (1) ρk − 2 < ρk − 1 < ρk; (2) ρk > ρk + 1. Among a set of ranks meeting these criteria, the optimal rank was then identified as the one at which CCC is maximized. The optimal rank selected for the pan-tissue atlas was 12 (Extended Data Fig. 3a,b).
Specifically correlated subset pairs
CoVarNet utilizes Pearson correlation coefficients to assess whether any two cell subsets co-occur. For a given set of s cell subsets, pairwise correlation tests are performed based on the frequency matrix, resulting in an s × s correlation coefficient matrix (denoted R). To quantify the specificity of correlations, an indicator is defined. For each element rij (i < j) in R, its background set Sij and specificity index Spec (rij) are defined as:
$$S_ij=\ k\ne i\\cup \ k\ne j\$$
$$\rmSpec(r_ij)=\frac S_ij$$
In other words, the specificity index is defined as the fraction of elements in the background set that do not exceed ri j. The specificity cutoff is determined by an automatic method. If n and N represent the assumed number of subsets in each CM and the total number of subsets, then the specificity cutoff Cutoff (n, N) will be determined as:
$$\rmC\rmu\rmt\rmo\rmf\rmf(n,N)=1-\frac(n-1)\times 2-1(N-1)\times 2-1$$
This approach enables a balanced assessment of the number of subsets within CMs and their co-occurrence. Specifically correlated subset pairs are determined jointly by the correlation (coefficient and FDR) and specificity. We generated 147 pairs for the pan-tissue atlas. These pairs were visualized as a global network (Supplementary Fig. 4).
Construction, evaluation and visualization of CM networks
For each NMF factor, the top subsets are designated as potential nodes, and edges connect specifically correlated subset pairs, removing isolated nodes. In each constructed CM network, the connectivity score is calculated as the ratio of observed edges to the total possible edges among all nodes within that network. The statistical significance of this score is assessed using a permutation test (n = 10,000) on the node labels. We used the igraph R package to visualize the CM networks, with nodes colour-coded by cell type and edge colour gradients scaled to reflect specificity.
CMT classifications of samples
The CM activities in individual samples are measured by the coefficient matrix from the NMF procedure, with the sum of activities for all CMs equalling 1 for each sample. Each sample was assigned a CMT label based on its most abundant CM. For instance, if a sample exhibited the highest activity of CM01 among all CMs, it was labelled as CMT01. All healthy single-cell samples used across tissues were stratified into 12 CMT groups (Extended Data Fig. 3e).
Integrative analysis of scRNA-seq and GTEx RNA-seq
We utilized GTEx24 RNA-seq datasets to validate the CMs defined by single-cell data (Extended Data Fig. 4).
RNA-seq data preprocessing
We retrieved gene transcripts per million (TPM) and metadata for 17,382 bulk RNA-seq samples from the GTEx Portal (V8 release)65. Samples derived from cell lines were excluded, and the ‘Cervix uteri’ category was merged into the ‘Uterus’ category for consistency. To ensure consistency, only tissues represented in the single-cell cohort were retained, narrowing down to a total of 12,240 samples spanning 23 tissues for further analysis. Gene expression data were re-normalized to a uniform library size of 10,000 and log-transformed for comparability with single-cell data.
CMT classifications of RNA-seq samples
We began by identifying DEGs among pseudo-bulk CMT samples. The top ten DEGs, ranked by fold change, were designated as CMT signature genes. Utilizing the Seurat R package66, we applied the AddModuleScore function to calculate scores for individual RNA-seq samples based on these CMT signature sets. All negative scores were adjusted to zero, and 2.3% (278 out of 12,240) of samples with the highest score less than 0.2 were excluded to ensure robust classification. Ultimately, the remaining 11,962 samples were categorized into 12 distinct CMTs, facilitating a detailed examination of CM representation across the analysed tissues.
Tissue prevalence of cell subsets and CMs
To assess the prevalence of cell subsets across tissues, we compared observed (o) to expected (e) cell numbers for each subset-tissue combination, expressed as Ro/e = observed/expected, following established methods35,38,39. Expected cell numbers for each subset–tissue combination were derived from the Chi-square test, with enrichment defined as Ro/e > 1 (Fig. 1e and Supplementary Fig. 2). For the assessment of each CM, we computed tissue-level CM activities by averaging its activity across all samples within each tissue. The Ro/e ratio indicated the tissue distribution of CM profiles (Fig. 2c). To compare CM enrichment across 23 overlapping tissues between bulk and single-cell analyses, we independently calculated Ro/e ratios for each data type and combined them for comparison (Extended Data Fig. 4f). Results were visualized using the ComplexHeatmap R package67.
Analysis of spatial transcriptomics data
Data collection
We gathered published spatially resolved transcriptomics datasets (Visium and Xenium) of various human tissues and cancer types. Detailed accession numbers and references for these datasets are provided (Supplementary Table 3).
Cell subset identification
For deconvoluting spatial transcriptomics data, we utilized cell2location25, a Bayesian model capable of accurately resolving fine-grained cell types within spatial data. Utilizing both healthy and cancer datasets, we used the corresponding integrated scRNA-seq data as a reference to obtain cell-type signatures. Prior to this process, the scRNA-seq data was subsampled to 1,000 cells per cell subset. In cases where cell types comprised fewer than 1,000 cells, all available cells were included. Following the recommended guidelines of cell2location, we set N = 5 as the expected cell abundance per spot and αy = 20 to regularize within-experiment variation in RNA detection sensitivity. The output yielded the expected cell abundance per cell subset in each spot.
CM activities
To quantify and visualize CMs in spatial transcriptomics, we aggregated the abundance of the component cell subsets within each CM, applying weights derived from NMF results. The resulting CM activities were then scaled to a uniform standard, with the 99th percentile value across all CMs being set to 1, allowing for a direct comparison of their relative magnitudes.
Colocalization scores
To assess the colocalization of cell-subset components within CMs across Visium spots, we calculated the colocalization score for individual spatial sections. For each CM, we calculated Spearman correlation coefficients between subset pairs where at least one subset is within this CM, resulting in a set of correlation coefficients denoted as S. The median correlation coefficient within the CM is termed r. The colocalization score for each CM is defined as the proportion of correlations in S that are less than or equal to r, providing a measure of colocalization relative to global contexts.
Aggregation scores
To assess the regional aggregation of cell-subset components within CMs in spatial transcriptomics, we utilized the global bivariate Moran’s I68 using the spdep R package. Similar to the colocalization score, the aggregation scores were calculated using global Moran’s I instead of the correlation coefficient.
Cellular niches
To provide orthogonal validation for the identified CMs, we used CellCharter26 to identify cellular niches, clustering Visium spots based on both gene expression and spatial information to enable spatially informed niche categorization. This analysis was performed for each sample independently to mitigate batch effects, allowing cross-validation of results across samples from the same tissues.
Xenium analysis
The Xenium platform enables in situ characterization of hundreds to thousands of genes in cells and tissues with ultra-precise single-cell spatial imaging. Using published intestinal Xenium data, we characterized the spatial locations of CM02 and CM03. We first designed a gene panel to distinguish multiple cell subsets within these CMs, with gene transcript density serving as a proxy for CM intensity (Extended Data Fig. 5i). Spatial regions of the tissue (epithelium or lamina propria) were identified on the basis of k-means clustering (k = 2) in the original dataset. To assess CM distribution across tissue sub-regions, we selected six different areas within the intestinal mucosa and measured CM intensities, providing a spatially resolved, single cell-resolution distribution of CMs.
Cell–cell communication analysis
To disentangle complex cellular crosstalk within and across CMs, we performed ligand–receptor-mediated cell–cell communication analysis using single-cell data with the CellPhoneDB Python package11,27.
Global analysis agnostic to CMs
Considering the extensive number of cells, we conducted subsampling to equalize the contribution of each cell subset. Specifically, we subsampled the number of cells to 1,000 cells for each cell subset. However, if the total cell count for certain subsets did not exceed 1,000, all cells were included in the analysis. This approach aimed to ensure that the null distribution accurately represented all cell subsets, avoiding bias towards cell subsets with larger cell numbers. Subsequently, the CellPhoneDB procedure was used for statistical inference of cell–cell interaction specificity, allowing the derivation of cell–cell interaction counts among different cell types (Extended Data Fig. 6c) or CMs (Extended Data Fig. 6d) by averaging the results of corresponding cell subsets. We also validated these results using an alternative tool, CellChat69 (Extended Data Fig. 6e).
Comparative analysis informed by CMs
To explore the effect of tissue microenvironments (CMs) on cellular crosstalk within CMs, we conducted CellPhoneDB analysis using two groups of samples with high or low CM activities, respectively. For each CM, the ‘High’ group included all samples in which that CM showed the highest activity among all CMs, while other samples were used as the ‘Low’ group. Following the previously defined CMTs in ‘CMT classifications of samples’, taking CM01 as an example, all samples labelled as CMT01 were used as the High group, while other samples were used as the Low group. Subsequently, comparative analysis between the two groups focused on cell subsets that were components of each CM (Fig. 3g).
Cytokine response analysis
The recently reported Immune Dictionary provides a comprehensive overview of cell-type-specific responses to 86 cytokines28. Leveraging this foundational knowledge, we explored intercellular crosstalk within CMs through cytokine responses. CM07 and CM10 were excluded as they lack immune cell subsets, while CM12 had no significant cytokine outputs.
CM-dependent DEGs of cell subsets
The tissue microenvironments exerted a broad influence on cell-subset phenotypes in a CM-dependent manner. Specifically, for each immune subset, we identified its DEGs (log2-transformed fold changes >log2(1.2), FDR < 0.05, Student’s t-test) in samples from corresponding CMTs when compared with those from other CMTs, utilizing the Scanpy toolkit. These identified DEGs were interpreted as responses to cytokines within the tissue microenvironments associated with CMs.
Cytokine response inference
First, we built a database of cytokine signatures for each cell type, using supplementary table 3 from the Immune Dictionary publication28. Of note, we performed one-to-one conversion of mouse and human ortholog genes based on the Mouse Genome Informatics (MGI) database70 (http://www.informatics.jax.org/downloads/reports/HMD_HumanPhenotype.rpt). Subsequently, we conducted cell-type-aware immune response enrichment analysis using the hypergeometric test (FDR < 0.05) through the enricher function in the clusterProfiler R package71.
CM-specific cytokine networks
To visualize cytokine-mediated multicellular regulation, we constructed cytokine networks by considering both cytokine production and response in a CM-specific manner. For cytokines to be considered in a CM, the normalized expression value of the cytokine gene needed to exceed 0.1 in at least one of non-responsive cell subsets. In the case of heteromeric cytokines or cytokine complexes with two subunits, each subunit was separately represented. The igraph R package was used to generate visual representations of the cytokine networks.
Association analysis of CM activity and phenotypic factors
Tissues
We first examined the association between CM activities and tissues, excluding tissues with fewer than five samples. For each CM, we fitted a linear model, with adjusted R2 indicating the proportion of variance explained. The FDR of the F test was reported to ensure the robustness of the results. Given the strong tissue preferences of CMs, subsequent analyses focused on tissue-specific associations.
Sexes and age groups
In non-reproductive tissues, CM activities were compared between samples from male and female participants, with FDR-adjusted significance. Samples lacking age information were excluded from the analysis. Age data were categorized into groups: <35, 35–39, 40–49, 50–59, 60–69 and 70–85 years. For the breast dataset (D03) with over 100 samples (Supplementary Table 1), age was categorized as <50 or ≥50 years. Associations between CM activities and age groups were assessed, with adjustments for statistical significance. Specifically, we also examined associations between immune cell-subset frequencies and age groups in the spleen.
Additional phenotypes
We performed further association analyses between CMs and specific phenotypic factors. We analysed CM09 in relation to alcohol consumption in the lymph node, CM06 and CM11 with childhood tuberculosis in the lung, CM12 with menopause in the breast, and CM07 with menstrual cycle phases in the uterus.
CM05 regulators in the spleen
Inferring regulons
We used the pySCENIC30,72 pipeline to infer regulons for the four subsets (B03, B05, CD4T03, and I06) within CM05, performing the analysis separately for the three cellular lineages (B cells, CD4+ T cells, and innate lymphoid cells). For each subset, regulons were ranked on the basis of their regulon specificity scores (RSS), and the top 50 regulons with the highest RSS were selected for each cell subset. Seventeen regulons were shared among the four subsets.
Activities and target genes of shared regulons
Sample-level activities for shared regulons were determined by averaging cellular activities across all cells in each sample. The mean activity of each regulon was then compared across age-stratified sample groups. Target genes of shared regulons were compared across lineages, and a regulatory network was constructed to illustrate their interrelationships.
MCP analysis
MCP identifications
As different cell types within the same CMs tend to be exposed to similar tissue microenvironments, we hypothesized that they might exhibit coordinated responses. To investigate this, we utilized a method called DIALOGUE3 to map MCPs for CMs. This procedure involved setting the parameter (k = 3) and assessing the association between the MCPs identified and other phenotypes. This analysis was applied to CM12 in the breast and CM08, and resulted MCPs were termed as CM12 programme and CM08 programme.
Comparison between CM08 programme and other signatures
To compare CM08 programme and inflammatory and cytotoxic signatures, we calculated their overall expression in external RNA-seq data, as described previously73,74. Specifically, RNA-seq datasets of samples from individuals with advanced melanoma following anti-PD-1 therapies75 were downloaded from https://github.com/ParkerICI/MORRISON-1-public. The inflammatory signature genes of immune cells are defined as CD3D, IDO1, CIITA, CD3E, CCL5, GZMK, CD2, HLA-DRA, CXCL13, IL2RG, NKG7, HLA-E, CXCR6, LAG3, TAGAP, CXCL10, STAT1 and GZMB76. The cytotoxic signature genes are defined as IFNG, GZMA, GZMB, PRF1, GZMK, ZAP70, GNLY, FASLG, TBX21, EOMES, CD8A, CD8B, CXCL9, CXCL10, CXCL11, CX3CL1, CCL3, CCL4, CX3CR1, CCL5, CXCR3, NKG7, CD160, CD244, NCR1, KLRC2, KLRK1, CD226, GZMH, ITK, CD3D, CD3E, CD3G, TRAC, TRBC1, TRBC2, CD28, CD5, KIRDL4, FGFBP2, KLRF1, SH2D1B and NCR3 (ref. 77).
Inflammatory scores of CM12 subsets in the breast
We calculated the sample-level inflammatory scores for immune and fibroblast subsets within CM12. Fibroblasts and immune subsets were scored using the corresponding inflammatory gene sets. The inflammatory signature genes of fibroblasts are defined as PLAU, CHI3L1, MMP3, IL1R1, IL13RA2, TNFSF11, MMP10, OSMR, IL11, STRA6, FAP, WNT2, TWIST1 and IL24 (ref. 78). The inflammatory signature genes of immune cells are defined as above. Specifically, we first calculated the average gene expression among all cells of individual subsets for each sample. Subsequently, we used the R package AUCell30 to calculate the sample-level inflammatory scores.
Menopausal trajectory analysis
Discovery cohort
We constructed a menopausal trajectory in the breast (dataset D03; Supplementary Table 1) based on the frequencies of cell subsets within CM12, following the methodology described in a recent study5. To mitigate the impact of frequency differences between cell subsets, we applied z-score normalization to correct the frequency matrix. Subsequently, we computed the k-neighbourhood and performed clustering for breast samples using the function scanpy.pp.neighbours and scanpy.tl.leiden with default parameters. To model trajectories along menopause, we performed PHATE79 with a = 40, followed by pseudotime analysis using the Palantir80 standard pipeline. The starting point was defined as the cluster with a high proportion of pre-menopausal samples.
Validation cohort
The Reed dataset81 was used to validate the menopausal trajectory. For the epithelial-enriched (‘organoid’) samples in the dataset, cell subset annotation was performed with CellTypist7, using previously annotated gene expression profiles of breast tissue as a reference. The cell subset frequency matrix was subsequently input into the trajectory analysis as described above.
Pan-cancer single-cell atlas
To disentangle the rewiring of CMs along malignant progression, we constructed a pan-cancer transcriptomic atlas at single-cell resolution (Fig. 5a).
Data collection and preprocessing
Following the criteria set for healthy datasets, we selectively included scRNA-seq datasets from fresh (not frozen) samples without cell-type enrichment, generated using 10x Genomics single-cell (not single-nucleus) platforms. One exception is the ESCC_GSE160269 cohort, where samples represent a mixture of immune, epithelial, endothelial, and stromal compartments. Quality control and other preprocessing procedures were also applied consistently with healthy datasets. In total, more than 1,000 samples from 48 datasets were incorporated, collectively forming a cell atlas across 29 human major cancer types (Extended Data Fig. 10 and Supplementary Table 7).
Data integration, cell clustering and initial annotation
Following the methodology applied to healthy datasets, we performed dataset integration using BBKNN. Unsupervised clustering for all cells was carried out using the scanpy.tl.leiden function with a resolution of 0.1. Subsequently, we identified the eight broad cell types based on canonical markers.
Supervised cell annotation
To accurately determine cell identities in cancerous samples, we utilized a transfer-learning-based strategy for cell subset annotation. Initially, we curated a single-cell reference dataset that encompassed 76 non-epithelial cell subsets identified in healthy data and 15 cancer-associated subsets identified in various cancer types35,36,37,38,39 (Supplementary Table 9). The number of cells of each subset was subsampled to 1,000, unless the total cell count did not exceed 1,000. Subsequently, a transformer-based reference model was trained on the reference dataset. Following this, non-epithelial cells from the pan-cancer atlas were annotated using the reference model. These procedures were executed using TOSICA, a multi-head self-attention model, enabling interpretable cell-type annotation82. The epoch number for prediction was selected as 15 (Supplementary Fig. 8c). Cells with predicted probabilities <0.5 were removed (Supplementary Fig. 8d). In the end, a total of 3,038,535 high-quality cells from 1,062 samples of 717 donors were well annotated to be 91 non-epithelial subsets.
Rewiring of multicellular ecosystems in cancer
Samples used
To compare multicellular dynamics during tumour progression, we included eight cancer types that had at least three samples from each condition (healthy, adjacent non-tumour and tumour).
Interrogation of healthy CMs
To quantify healthy CMs across different conditions, we aggregated the abundance of the component cell subsets within each CM, applying weights derived from NMF results. The resulting CM activities were then rescaled to range from 0 to 1. For each cancer type, only the most dominant CM was considered.
Co-occurrence of cell subsets in individual cancer types
For each cancer type, we derived all specifically correlated cell-subset pairs using the correlation analysis module of CoVarNet. Compared with pan-tissue or pan-cancer analysis, the following more stringent cutoffs are used: coefficients >0.5, FDR <0.05 and specificity >0.95. Only identified specifically correlated subset pairs are used to perform comparison across eight cancer types.
Identification of cCMs using CoVarNet
Specifically, we generated a frequency matrix consisting of 91 subsets and 955 samples for the pan-cancer atlas. Specifically, the top 15 subsets of each factor were used as co-occurring node candidates in a single CM network for the pan-cancer analysis.
Interrogation of cCM02
To quantify cCMs across different conditions, we measured cCM02 activities using the same procedure as in healthy CMs.
cCM02 analysis
Co-occurring network
To explore the dynamics of cCM02 during tumour progression, we constructed two co-occurring networks of which nodes are the same as original nodes in cCM02, while edges were recalculated in two scenarios. One used healthy and adjacent non-tumour samples, while the other used tumour and adjacent non-tumour samples. Thus, CM networks were constructed using unaltered nodes and new edges.
Cytokine analysis
For each cell-subset component of cCM02, we identified its DEGs (log2-transformed fold changes >log2(1.2), FDR < 0.05, Student’s t-test) in tumour samples compared with adjacent non-tumour samples. These identified DEGs were used to perform cytokine analysis as described above.
MCP analysis
We conducted MCP identification using only tumour and adjacent non-tumour samples. Overall expression of MCP were calculated for the RNA-seq datasets from TCGA portal and microarray data of pre-invasive lung lesions45. TCGA datasets were downloaded using the TCGAbiolinks83 R package. Only projects with more than ten tumour or adjacent non-tumour samples were included. Datasets of pre-invasive lung lesions were downloaded from https://github.com/ucl-respiratory/preinvasive.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.