Laboratory animals
Cas9-expressing mice (Cas9 mice) were obtained from the Jackson Laboratory (https://www.jax.org/strain/024858). These mice (background C57BL/6N) constitutively express the Cas9 endonuclease and an eGFP reporter under the control of a CAG promoter knocked into the Rosa26 locus49. All screens in this study were performed with the Cas9 mice, including all NSC primary cultures and all in vivo work. We maintained a colony of Cas9 mice ranging in ages up to 28âmonths at the Stanford Comparative Medicine Building and the Neuroscience-ChemH building vivarium. As a negative control for the in vivo screens, male C57BL/6 mice obtained from the National Institute on Aging Aged Rodent colony were used at 18â21âmonths old. These mice were habituated in the Stanford facility for at least 2âweeks before initiation of experiments. Mice were maintained under the care of the Veterinary Service Center at Stanford University under IACUC protocols 8661.
Primary cultures of NSCs from young and old brains and activation experiments
For all experiments involving primary culture of NSCs, we pooled SVZs from pairs of male and female Cas9 mice, either 3â4âmonths old (young) or 18â21âmonths old (old). To generate primary cultures of NSCs from young and old mice, we microdissected SVZs into a small drop of PIPES buffer (pHâ7.4), minced them in a 10âcm tissue culture dish with about 100 chops of a scalpel blade and suspended the tissue in PIPES buffer before centrifugation for 5âmin at 300g, at which point the excess PIPES buffer was poured out. The pellet of minced SVZs was then enzymatically dissociated (in 5âml per 2 SVZs) with a mixture of HBSS (Corning, 21-021-CVR) with 1% penicillinâstreptomycinâglutamine (Gibco, 10378-016), 1âUâmlâ1 DispaseâII (StemCell Technologies, 07913), 2.5âUâmlâ1 papain (Worthington Biochemical, LS003126) and 250âUâmlâ1 DNAse I (D4527, Sigma-Aldrich), vortexed briefly and incubated at 37â°C for 40âmin on a rotator. The samples were then centrifuged at 300g for 5âmin at room temperature and resuspended in Neurobasal A medium (Gibco, 10888-022) with 1% penicillinâstreptomycinâglutamine (Gibco, 10378-016) and 2% B27 minus vitaminâA (Gibco, 12587-010) and triturated about 20 times, centrifuged and resuspended in complete âaNSC mediumâ, comprising NeurobasalâA (Gibco, 10888-022) supplemented with 2% B27 minus vitaminâA (Gibco, 12587-010), 1% penicillinâstreptomycinâglutamine (Gibco, 10378-016), 20ângâmlâ1 EGF (Peprotech, AF-100-15) and 20ângâmlâ1 bFGF (Peprotech, 100-18B) and placed in a humidified incubator at 37â°C and 5% CO2. After 3â4âdays, neurospheres emerged in the medium and were passaged by dissociation with 1âml Accutase (StemCell Technologies, 07920) for 5âmin at 37â°C, washed once with PBS and resuspended in aNSC medium. Neurosphere cultures were maintained with passaging every 2â3âdays, and all experiments were performed in cultures of fewer than 10 passages. Details on passage numbers are provided in experimental sections below. For cultures of qNSCs, the aNSC culture medium was changed to remove EGF and to add BMP4 (50ângâmlâ1) (Peprotech, 315-27). The Complete âqNSC mediumâ comprised Neurobasal-A (Gibco, 10888-022) supplemented with 2% B27 minus vitaminâA (Gibco, 12587-010), 1à penicillinâstreptomycinâglutamine (Gibco, 10378-016), 50ângâmlâ1 BMP4 (BioLegend, 94073) and 20ângâmlâ1 bFGF (Peprotech, 100-18B). To induce quiescence, tissue culture plates were pre-treated with PBS (Fisher Scientific, MT21040cv) containing 50ângâmlâ1 poly-d-lysine (PDL; Sigma-Aldrich, P6407) for 1âh and then washed 3âtimes with PBS before plating cells on plates in qNSC medium. The density of cells plated is important for induction of quiescence and the ability of qNSCs to reactivate, especially in the context of lentiviral infection. In optimizing the qNSC activation protocol, we observed that qNSCs seeded at the following densities were best for quiescence and activation experiments: 2âÃâ107 cells for a 15âcm plate; 1âÃâ106 cells per well for a 6-well plate; 2âÃâ105 cells per well for a 24-well plate; and 1âÃâ105 cells per well for a 96-well plate. For activation of qNSC cultures, cells were washed once with PBS, and then aNSC medium was added to the plate and refreshed every 2âdays. For plating, cells were manually counted with a haemocytometer or using a Countess II FL Automated Cell Counter (Life Technologies, AMQAF1000). NSC primary cultures were tested quarterly for the presence of mycoplasma and tested negative.
Tissue culture plastics
We found that primary cultures of NSCs were sensitive to the tissue culture plastic products used. Specifically, passaging NSCs in conical tubes manufactured by Genesee (15âml conical tubes, 28-103) resulted in death of the NSC cultures within 1âweek of brief exposure to the plastic during passaging. Plastics from the following manufacturers were assessed to be suitable for NSC growth both in detached and adherent conditions: Thermo Fisher 15/50âml Falcon tubes (14-959-53A/14-432-22), 15âcm, 10âcm, 6-well, 12-well, 24-well and 96-well Falcon tissue culture dishes (353025, 08772E, 08-772-1B, 08-772-29, 08-772-1, 087722c, respectively).
Lentivirus production
Genome-wide virus library preparation
For lentiviral production, we used human embryonic kidney 293T cells. 293T cells were from the American Type Culture Collection (they were not authenticated). They were tested for the presence of mycoplasma in a quarterly manner and tested negative. 293T cells were seeded in DMEMâ+â10% FBS (Gibco 10099141)â+â1à penicillinâstreptomycinâglutamine (Gibco, 10378-016) at a density of 2âÃâ107 cells in 15âcm plates. One day later, 293T medium was replaced with 18âml fresh medium and the cells were transfected using the polyethylenimine (PEI) (1âmgâmlâ1, Polysciences, 23966-2) transfection method, mixing plasmids as follows: 2.27âμg each of third-generation lentivirus packaging vectors pMDLg, pRSV and pVSVG (obtained from the laboratory of M.âBassik), along with 45âμg of the pooled sgRNA genome-wide plasmid library (https://www.addgene.org/pooled-library/bassik-mouse-crispr-knockout/). The sgRNA library targets around 23,000 protein-coding genes in the genome, with 10 unique sgRNAs per gene, and15,000 control sgRNAs (about 245,000 sgRNAs in total)29. The sgRNA plasmid library, consisting of 20 sublibraries, was mixed proportionally to the number of sgRNAs in each library. One day after PEI transfection, the medium was changed to 18âml of NeurobasalâAâ+â1à penicillinâstreptomycinâglutamine (Gibco, 10378-016). After 1âday, the viral containing supernatant was collected on ice and stored at 4â°C. Fresh medium was added to the 293T cells and collected again after 24âh and again at 48âh (a total of 3 collections of 18âml of virus supernatant). All 3âsupernatants were combined, filtered through a 0.45âμm filter (Stericup, EMD Millipore, S2HVU02RE) and frozen at â80â°C in 10âml aliquots in 15âml conical tubes. For plasmid library re-amplifications, we electroporated 1âμl of 25ângâμlâ1 of each library into 50âμl bacteria (Lucigen, 60242-2), with 1.8âkV, 600âΩ and 10âμF in a 0.1âcm cuvette (Gene Pulser Xcell, Bio-Rad, 1652662). After electroporation, we allowed bacteria to recover in Lucigen recovery medium for 2âh in 15âml conical tubes, shaking at 37â°C. We plated 1âμl of the transformation onto a LBâ+âcarbenicillin (100âμgâmlâ1, Sigma-Aldrich, C9231-1G) agar plate to confirm transformation efficiency, and the remainder of the recovery suspension was placed into 0.5âlitre LBâ+âcarbenicillin (100âμgâmlâ1) liquid medium in a 2âlitre flask for 16âh of shaking at 37â°C, and DNA was purified using a Maxiprep kit (Thermo Fisher Scientific, FERK0492) according to the manufacturerâs protocol.
sgRNA sublibrary design
We designed five sublibraries of sgRNAs to test gene hits from the in vitro screens in the brain in vivo (Figs. 1 and 2). Our selection criteria were as follows. For the topâ10 gene list, we selected all significantly enriched genes (FDRâ<â0.1) from the first 2 in vitro genome-wide screens, selecting any gene that was significant in both screensâ1 and 2, at any time point, dayâ4 or dayâ14 (for example, a screenâ1 dayâ4 hit and an overlapping screenâ2 dayâ14 hit would be added to the list). With that list, we ranked the genes based on the CasTLE gene score average from both screens and both time points (that is, average of all: screenâ1 dayâ4 or dayâ14, screenâ2 dayâ4 or dayâ14). This library was selected based on the first two in vitro genome-wide screens only, because at the time of library design, only the first two in vitro screens had been completed. Similarly, the âdepletedâ gene list was also selected based on the first two screens. We selected all significantly depleted genes (FDRâ<â0.1) from the first 2 in vitro genome-wide screens, selecting any gene that was significant in both screensâ1 and 2, at any time point, dayâ4 or dayâ14 (for example, a screenâ1 dayâ4 hit and an overlapping screenâ2 dayâ14 hit would be added to the list). With that list, we then removed any gene that was significantly (FDRâ<â0.1) depleted in qNSCs of screenâ1 or 2 of any age. The final list was then selected by removing unannotated genes (for example, GM3264 and GM3164) and focusing on genes with associated publications. For the âglucose uptake/human diseaseâ list, we selected genes that were significantly (FDRâ<â0.1) enriched in 2 out of 3 in vitro genome-wide screens, at dayâ4 or dayâ14 (for example, a screenâ1 dayâ4 hit and an overlapping screenâ2 dayâ14 hit would be added to the list). The list of enriched genes was analysed by GO term analysis (see the section âComputational analysis of CRISPR screensâ). From the GO Molecular Function (2018) database, the d-glucose transmembrane transporter activity (GO:0055056) and sugar:proton symporter activity (GO:0005351) terms were both in the topâ10, with genes Slc2a4, Slc2a12 and Slc45a4. The other genes in the âglucose uptake/human diseaseâ list were selected based on one of two criteria: (1) genes implicated in human disease: Snrpb2 (Alzheimerâs disease)85, Sorl1 (Alzheimerâs disease)86 and C1qtnf5 (human ageing)87; or (2) genes that are significantly (Pâ<â0.05) upregulated in qNSCs in old mice: Slit2 (refs. 25,88), Ier2 (ref. 25), Cdkn1a25,88 and Ecscr25. For the âcytoplasmic ribonucleoprotein granulesâ library, in the GO term analysis of gene knockouts that boosted old NSC activation, the terms P-body (GO:0000932), cytoplasmic ribonucleoprotein granule (GO:0036464), ribonucleoprotein granule (GO:0035770) and cytoplasmic stress granule (GO:0010494) all came up in the list, although most were not significant. From this, we proposed that cytoplasmic granule structures could impede old NSC activation. We took the entire GO term cytoplasmic ribonucleoprotein granule (GO:0036464) and selected gene knockouts that had the greatest difference in effect between young and old NSC screens. Many of these genes did not demonstrate any significant effect in our in vitro screens, other than Dis3l2, Edc3 and Mbnl1, which all significantly boosted old NSC activation in at least two out of three screens. The final list of genes was the âpublished NSC regulatorsâ list, which we chose based on searching the literature for genes that had previously been implicated in regulation of NSC function and behaviour. We did not select based on functional effect prediction.
sgRNA plasmid sublibrary cloning for in vivo screens
The sgRNA expressing plasmid MCB320 (https://www.addgene.org/89359/) was digested with the BlpI and BstXI restriction enzymes, the band was gel-extracted and purified and used for a pooled ligation reaction. We selected five sgRNAs from each gene of interest, based on the five out of ten sgRNAs most enriched or depleted in our genome-wide in vitro screen. For the forward oligonucleotide of each sgRNA sequence, we added the following sequences: 5â²-ttgg and 3â²-gtttaagagc. For the reverse complement oligonucleotide of each sgRNA sequence, we took the reverse complement of the sgRNA sequence and added 5â²-ttagctcttaaac and 3â²-ccaacaag. To clone a pool of 10 genes, we selected the 50 sgRNAs pairs targeting the 10 genes and annealed the sgRNA pairs in separate annealing reactions in a 100âμl of IDT duplex buffer (11-05-01-12) with 1âµM forward and reverse oligonucleotides. We incubated the oligonucleotide pairs at 95â°C for 5âmin and then allowed the oligonucleotides to gradually anneal at room temperature. We mixed all 50 annealed oligonucleotide pairs into one pool, diluted it 1:20 in IDT duplex buffer and then used 1âμl of annealed oligonucleotide pool in a ligation reaction with 500âng of digested MCB320 backbone. We used 1.5âμl of the ligation mix and electroporated 30âμl competent bacteria (Lucigen, 60242-2) with 1.8âkV, 600âΩ and 10âμF in a 0.1âcm cuvette (Gene Pulser Xcell, Bio-Rad, 1652662). We plated the entire recovered transformed bacteria on a 10âcm LBâ+âampicillin (100âμgâmlâ1, Sigma-Aldrich, A9518-100G) plate, allowed overnight recovery, and the next day added 5âml LB to the bacterial lawn and scraped it with a sterile silicon scraper. The resuspended bacterial mix was transferred to a clean collection tube, the plate was again rinsed with an additional 5âml LB and transferred to same tube for overnight growth in 500âml LBâ+âampicillin (100âμgâmlâ1) for Maxiprep (Thermo Fisher Scientific, FERK0492) according to the manufacturerâs protocol. For library re-amplification, we performed the same transformation and amplification procedure.
Concentration of virus for in vivo and in vitro subscreens
For in vivo and in vitro subscreens, we generated virus the same way as our genome-wide virus libraries but with modifications. We plated four 15âcm plates of 293T cells for a total of 200âml of collected virus after 3âdays of collecting at 4â°C, but rather than directly freezing the virus, we performed ultracentrifugation to concentrate the virus. For ultracentrifugation, we sterilized 30âml ultraclear tubes (Beckman Coulter 344058) under UV (TC room biosafety cabinet) for 15âmin. We then put the tubes on ice, allowed 15âmin to cool and then added 30âml of virus and centrifuged at 16,500âr.p.m. for 1âh at 4â°C. We carefully decanted the supernatant using serological pipettes, leaving 1âml medium in the bottom of the tube, adding 30âml more virus-containing medium and centrifuging again. We repeated the decanting, refilling and centrifugation of the same tube, concentrating a total of 180âml of virus supernatant into a single tube. After the last ultracentrifugation, we removed most of the supernatant with a serological pipette, and the last 1âml with a P1000 pipet tip from the side of the tilted tube, so as not to disturb the viral pellet. The viral pellet was usually visible in centre of all of the tubes. We resuspend in 60âμl ice-cold PBS (1/3,000th original volume) by pipetting up and down about 60 times, being careful not to produce air bubbles. The concentrated resuspended virus was then aliquoted into PCR strip tubes in 5âμl aliquots and placed onto dry ice. After 15âmin, the virus was transferred to â80â°C for storage. For experiments, virus was thawed on ice and injected into the brain or added to cell culture within 30âmin of thawing. We assessed virus infectivity of each batch by performing serial dilution (3âμl, 1âμl, 0.5âμl) infections of 2âÃâ105 293T cells in 24-well culture plates for 16âh of infection and then performing FACS analysis 48âh later to detect the perâcent of cells expressing the mCherry reporter. For each experiment, we normalized virus infectivity (viral titre) across treatments by adjusting the concentrations of virus added in PBS.
Genome-wide knockout screens in primary cultures of NSCs
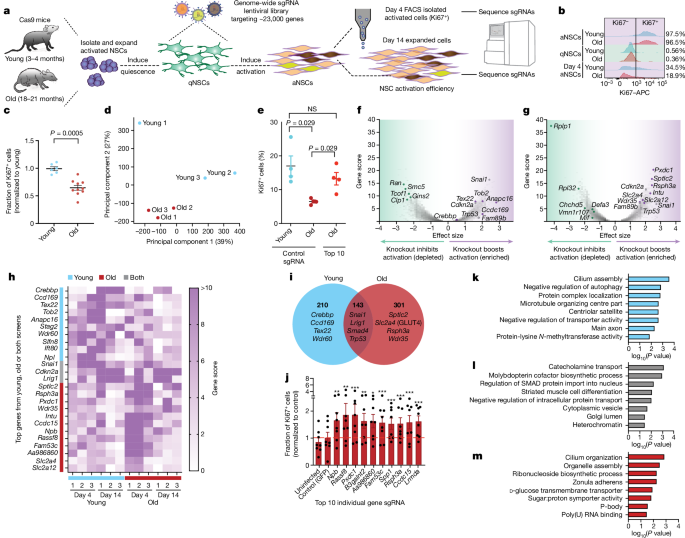
For each genome-wide screen, primary cultures of NSCs derived from a pool of three male and three female Cas9 mice were used for each independent biological replicate. In total, three independent genome-wide screens, each performed with independent young and old NSC pooled from six mice, were conducted. For each independent screen, young and old NSC cultures were processed in parallel at each stage of sample processing. The young and old NSC culture passage numbers were kept the same and at the start of screen were as follows: screenâ1, passageâ8; screenâ2, passageâ7; screenâ3, passageâ12. To measure the growth rates of young and old cells at each passage, we counted cells using a Countess 3 cell counter (Thermo Fisher, A50298). The young and old cells showed comparable growth rates (Extended Data Fig. 1c). To expand the NSCs up to 1.4âÃâ109 cells (the equivalent of 140 15âcm plates) required for each biological replicate, 1âÃâ107 NSCs were passaged and expanded into 15âcm plates every 2â3âdays, with feedings every 2âdays (alternating between doubling the medium (with 2à growth factor aNSC medium) or complete medium exchange). For each screen, 70 plates of 2âÃâ107 qNSCs were seeded at dayâ0 (see below for library coverage calculations). After 4âdays in qNSC medium, the cells were incubated with the genome-wide sgRNA lentivirus library (see above). For this, the sgRNA lentivirus library was freshly thawed at room temperature and diluted 1:5 in Neurobasal medium and then B27 and growth factors were added to make it qNSC medium, and 18âml of this mix was added to plates for 16âh of overnight infection. The virus dilution added was based on viral titration experiments determined to achieve about 30% culture infection of cells to ensure each cell only received a single sgRNA. Therefore, infecting the starting 1.4âÃâ109 cells at 30% infection would result in 4.2âÃâ108 infected cells, giving us a coverage of about 1,700 cells per sgRNA (around 243,000 total sgRNAs). Note that these numbers represent the starting library coverage, but the cells do expand over the course of activation; therefore, the final cell numbers at end of experiment are orders of magnitude larger. The infected cells were then left in qNSC medium for an additional 5âdays before transition to aNSC medium for activation. After 4âdays of activation, the cells were dissociated using Accutase (Stem Cell Technologies, 07920) for 15â30âmin at 37â°C (until most cells rounded up) and gently scraped with silicone cell scrapers (Fisher Scientific, 07-200-364) and split into 2 groups: 55 plates of NSCs were processed for the dayâ4 Ki67 FACS sorting (see below), and the other 15 plates of cells were placed into aNSC culture for 10âdays of further expansion as neurospheres (dayâ14 timepoint). The dayâ4 FACS-sorted young and old cells were sorted to have equal numbers of Ki67+ cells from both ages for each screen for downstream analysis. See the section âIntracellular FACSâ below for dayâ4 FACS protocol. The final number of sorted cells for each age in each screen was as follows: screenâ1 had 2.2âÃâ107 sorted Ki67+ cells; screenâ2 had 1.41âÃâ107 Ki67+ cells; and screenâ3 had 1âÃâ108 Ki67+ cells. After sorting, the methanol-fixed cells were centrifuged at 700g for 5âmin, the supernatant FACS buffer was decanted and the cell pellets were frozen at â80â°C until genomic DNA extraction. To extract genomic DNA of sorted and methanol-fixed cells, the cell pellets were defrosted at room temperature and then processed by resuspending in 5âml of TE 1% SDS (Thermo Fisher Scientific, 15525017) and incubated at 65â°C for 16âh. The cell suspension was then treated with 50âμl proteinaseâK (Fisher Scientific, 25-530-049)(20âmgâmlâ1) for 2âh at 37â°C. Samples were processed for genomic DNA extraction using Zymo Research ChIP DNA clean and concentrator (Zymo, D5205) according to manufacturerâs protocol. The dayâ14 expanding neurospheres were immediately centrifuged at 300g for 5âmin and then processed for genomic DNA extraction with a Qiagen QiaAmp DNA Blood Maxi kit (51194), adding 5âÃâ107 cells per column and according to the manufacturerâs protocol.
sgRNA PCR amplification and sequencing
After genomic DNA isolation, sgRNA was amplified from the genome in two successive, nested PCR reactions. For the nested PCR reactions, we used either Herculase II Fusion polymerase (Agilent, 600679) for screenâ1 or Q5 DNA polymerase (Fisher Scientific, M0491L) for screensâ2 and 3, and Q5 DNA polymerase for in vivo screens, according to the manufacturerâs protocol. In optimizing this PCR reaction, we found that Herculase II polymerase was outperformed by Q5 polymerase, with Q5 polymerase requiring fewer PCR cycles to obtain more amplicon product, which is why we switched to Q5 DNA polymerase. We used 5âμg genomic DNA in 50âμl reactions to run on a thermocycler. For the first PCR, we used primers MCB1562 (aggcttggatttctataacttcgtatagcatacattatac) and MCB1563 (acatgcatggcggtaatacggttatc) (1âµM final concentration), with PCR cycles as follows: 98â°C for 2âmin, 19 cycles of (98â°C for 30âs, 59.1â°C for 30âs, 72â°C for 45âs), followed by 72â°C for 3âmin. We pooled all the PCRâ1 cycle products and then used 5âµl of the pool in a second PCR reaction (PCRâ2) with the same conditions but using different primers: MCB1439 (caagcagaagacggcatacgagatgcacaaaaggaaactcaccct) and a barcoded primer (aatgatacggcgaccaccgagatctacacGATCGGAAGAGCACACGTCTGAACTCCAGTCACXXXXXXCGACTCGGTGCCACTTTTTC, where XXXXXX is the 6-digit barcode for high-throughput sequencing sample identification). The second PCR reaction was run for either 30âcycles (in vitro screenâ1 and in vivo screens) or 18 cycles (in vitro screensâ2 and 3). The resulting PCR products were all resolved on a 1.5% DNA agarose gel. The 272âbp band was extracted (Qiaquick Gel extractions kit, 28706), eluted in 10âμl ultrapure water (Invitrogen, 10977023) and assessed on a bioanalyzer (Agilent, Bioanalyzer 2100). Final libraries were combined into a pool at equal concentrations for sequencing on an Illumina Novaseq S4 system (by Novogene for the genome-wide in vitro screen) or on an Illumina MiSeq system (by the Stanford Genomics Facility for in vivo screens), sequencing to a depth of about 1âÃâ107 or 5âÃâ105 reads per sample for in vitro and in vivo screens, respectively.
Computational analysis of CRISPRâCas9 screens
For both the in vitro and in vivo screens, our analyses were performed using the CasTLE pipeline28. All of the source scripts can be found at Bitbucket (https://bitbucket.org/dmorgens/castle/downloads/). In brief, for each screen, the raw screen fastq files were aligned to the sgRNA library sequence (mm-Cas9-10 or one of a custom 10 gene libraryâ+âcontrol sgRNAs) to make count files using the makeCounts script. The count files were then analysed using the analyzeCounts CasTLE script, comparing each screen timepoint to the starting plasmid sgRNA library count file (in vitro screens) or the sequenced 24-h SVZ count file (in vivo screens), which we sequenced in parallel with screen libraries. We then calculated Pâvalues for all genes in each screen by running 100,000 (in vitro screens) or 10,000 (in vivo screens) permutations with the addPermutations CasTLE script. For each genome-wide screen, we corrected for multiple hypotheses on the around 23,000 gene associated Pâvalues using the Python Statsmodel module, with the BenjaminiâHochberg method, and classified genes as significant using a FDRâ<â0.1 cut-off. For the in vivo screens, we classified genes as hits if their CasTLE computed 95% confidence interval did not contain 0. The library diversity of each sample was displayed using the plotDist CasTLE script. The screen results and individual gene sgRNA enrichment plots were visualized using the plotVolcano and plotGene scripts, respectively. We note that in in vivo screens, there was a bimodal distribution of control sgRNAs, which is most likely due to the fact that a control sgRNA infected a NSC that was, at the time, highly actively proliferating and that will naturally enrich in the olfactory bulb.
Generation of gene lists for the genome-wide screens
To generate the final gene lists for the genome-wide screens, we used all genes that were significant (FDRâ<â0.1) in 2 or more independent screens (screensâ1 and 2, 2 and 3, or 1 and 3), at any time point (dayâ4 or dayâ14; for example, a screenâ1 dayâ4 hit and an overlapping screenâ2 dayâ14 hit, would be added to the list). For PCA, we used the Python sklearn.decomposition.PCA module with CasTLE-computed gene scores as input (Fig. 1d and Extended Data Fig. 1gâi). We performed gene set enrichment analysis by inputting gene lists into the EnrichR online portal (https://maayanlab.cloud/Enrichr/)89,90, and then focusing on the âOntologiesâ tab with GO Biological Process (2018), Molecular function (2018) and Cellular components (2018), sorting the terms based on Pâvalue, which is computed by EnrichR using the Fisher exact test.
Assessment of potential outliers in the genome-wide screens
To test for potential outliers, we compared CasTLE scores for all genes in the genome-wide screen between each replicate at dayâ14 for the young NSC screens (Extended Data Fig. 1dâf). Correlations between replicates were calculated using Spearmanâs correlation test. We also examined the principal component (PC) loadings of dayâ14 young and old in vitro genome-wide screens (PC1 and PC2) and dayâ4 young and old in vitro genome-wide screens (PC3 and PC4). PC loadings were extracted using the Python sklearn.decomposition.PCA module. We performed gene set enrichment analysis by inputting the top 50 gene knockouts contributing to the PC into EnrichR online portal (https://maayanlab.cloud/Enrichr/)89,90, and then focusing on the âOntologiesâ tab with GO Biological Process (2018), Molecular function (2018) and Cellular components (2018), sorting the terms based on Pâvalue, which is computed by EnrichR using the Fisher exact test. The PC loadings and the GO terms are included in Supplementary Table 4. The GO terms of the genes that contribute to the replicate youngâ1 at dayâ14 not clustering with the other young replicates are enriched for cytosolic proteasome complex (GO:0031597) and proteasome-activating ATPase activity (GO:0036402). As mentioned above in the section âGeneration of gene lists for the genome-wide screensâ, we chose hits from the screen that were significant in two or three replicates to avoid having one of the screens skew the data. We also investigated the loading of all PCs for both dayâ4 and dayâ14 and performed GO term analysis on the genes underlying all PCs (Supplementary Table 4). Genes and GO terms underlying the technical variance for dayâ4 samples (PC1, PC2 and PC4) are involved in cell division, proteostasis and transcription/translation. Thus, one possible source of variance in this in vitro NSC system could be due to lentiviral infection (affecting cell survival/cell proliferation) or bottlenecking during passaging.
Comparison with published screens and databases
We tested whether the genes significantly depleted at the dayâ14 timepoint overlapped with a list of common essential genes. We generated a list of dayâ14 significantly (FDRâ<â0.1) depleted genes in both the young and old screens (Supplementary Table 1). We identified the overlap between significantly depleted genes in the NSC screens with the Core Essential Genesâ2 list91 and the Online GEne Essentiality database (https://v3.ogee.info/#/home)92 (Extended Data Fig. 1l,m). Pâvalues were calculated using a Fisherâs exact test. There was a small but significant overlap between the known essential gene lists and the significantly depleted NSC gene list. Thus, these in vitro genome-wide screens captured essential genes that are shared with published datasets but also captured unique genes for which knockout affected cell survival or activation in NSCs.
Intracellular FACS for Ki67
For the genome-wide screen and for other qNSC activation experiments, we FACS-isolated proliferative cells (Ki67+) as follows. Cells were dissociated with Accutase (StemCell Technologies, 07920) for 5âmin, collected into conical tubes and centrifuged at 300g for 5âmin. Cells were resuspended in PBS at 5âÃâ107 cells in 1âml (or 1âÃâ105 cell in 100âμl), and then 9âml (or 900âμl) ice-cold 100% methanol was added and cells were agitated for 15âmin at 4â°C. Cells were then centrifuged at 500g for 5âmin and resuspended for a wash in 3âml PBS and centrifuged again at 500g for 5âmin. Cells were then resuspended in 3.5âml staining solution: Ki67-APC (eBioscience, 17-5698-82) 1:300 in PBS, 2% FBS (Gibco, 10099141) at 4â°C. Samples were agitated for 30âmin at room temperature in the dark, and then 10âml PBS was added before centrifugation at 700g for 5âmin. Samples were then resuspended (25âml per 5âÃâ107 cells) in FACS buffer: PBS, 2% FBS, DAPI (Fisher Scientific, 62248, 1âmgâmlâ1) 1:5,000. Each sample was filtered with FACS-strainer cap tubes (Fisher, 08-771-23), immediately before FACS sorting. Cells were sorted on an Aria BD FACS Aria with a 100âμm nozzle at 13âp.s.i., with BD FACSDiva software (v.8.0.1), and FlowJo (v.10) software was used for data analysis.
Assessment of NSC activation for in vitro subscreen and glucose intervention
For testing the topâ10 gene library (in vitro subscreen) (Fig. 1e) and glucose intervention (Fig. 4i), we performed qNSC activation experiments in a 24-well or 96-well format. We seeded 2âÃâ105 cells in a 24-well format or 1âÃâ105 cells in a 96-well format. After 4âdays in qNSC medium, with medium changes every 2âdays, concentrated virus was then added to the cells. We added 3âμl equal titre virus (see the section âConcentration of virus for in vivo and in vitro subscreensâ) to each 24-well containing 500âμl qNSC medium, or 0.1âμl virus to 100âμl in each 96-well experiment. We left the virus in medium with cells for 16âh, and then refreshed the medium. At 5â6âdays after infection, the cells were washed 1à in PBS and then either transitioned to aNSC medium for activation (Fig. 1e) or incubated with qNSC medium with or without glucose for 2âdays and then transitioned to aNSC medium for activation (Fig. 4i). aNSC medium was exchanged once after 48âh and Ki67 intracellular FACS was performed at dayâ4 after infection (see the section âIntracellular FACSâ above). Even though overall activation was decreased in lentivirally infected cells compared with non-infected cells, the difference in activation between old and young NSCs was preserved.
Assessment of the impact of individual gene knockout on NSC activation
To test the impact of individual gene knockouts on NSC activation (Fig. 1j), we used 8 independent NSC cultures from old (18â21âmonths old) mice, each culture being a mix of 1 male and 1 female mouse. We infected these NSC cultures with purified lentiviruses expressing five sgRNAs per gene. We evaluated the top 10 genes (screensâ1 and 2) (Supplementary Table 3). To assess the effect of each individual gene knockout on NSC activation, we seeded 3âÃâ105 NSCs in a 24-well format. After 4âdays in qNSC medium, with medium changes every 2âdays, qNSCs were incubated with fresh lentiviruses with equal titre. Lentiviruses were generated as described in the section âLentivirus productionâ using 293T cells. For these experiments, lentiviruses expressing sgRNAs to individual genes were collected by incubating 293T cells directly in qNSC medium. The supernatants were collected and their titres were tested, using serial dilutions to achieve a similar 50â70% range of infection of 293T cells. Supernatants were added to the qNSC wells for 16âh, and then the medium was changed to qNSC medium. Six days after infection, the cells were washed 1à in PBS and then transitioned to aNSC medium for activation. aNSC medium was exchanged once after 48âh and then Ki67 intracellular FACS was performed at dayâ3 after infection (see the section âIntracellular FACSâ above).
Validation of individual knockout efficiency
We validated the knockout efficiency for seven individual genes in qNSC cultures in two independent experiments:
For experimentâ1, young and old NSCs were seeded in a 24-well PDL pre-coated plate at a density of 2â3âÃâ105 cells per well and incubated in qNSC medium. After 4âdays with medium changes every 2âdays, qNSCs were infected with lentiviruses expressing sgRNAs targeting each gene (5 sgRNAs per gene) as described in the section âAssessment of the impact of individual gene knockout on NSC activationâ (see Source Data Extended Data Fig. 1n for sgRNA sequences). Six days after infection, cells were washed with PBS then lysed directly with DirectPCR Lysis reagent (Viagen Biotech, 102-T) with 1% ProteinaseâK (Fisher Scientific, 25-530-049) for 10âmin at room temperature. The supernatant was pipetted repeatedly, then transferred to PCR strip tubes and incubated at 65â°C for 25âmin, and then 95â°C for 15âmin in a thermocycler. We amplified genomic DNA with primer pairs surrounding the sgRNA-editing sites (see Source Data Extended Data Fig. 1n for amplification primers), using Q5 polymerase (Fisher Scientific, M0491L) and the following program: 30âs of annealing step at 55â°C and 1âmin of extending step at 72â°C for 40 cycles total.
For experimentâ2, we cloned 5 sgRNAs for each individual gene, using the same methodology as described in the section âsgRNA plasmid sublibrary cloning for in vivo screensâ above. For lentiviral production, 293T cells were seeded in DMEMâ+â10% FBS (Gibco 10099141)â+â1à penicillinâstreptomycinâglutamine (Gibco, 10378-016) at a density of 13âÃâ106 cells in 15âcm plates. One day later, 293T medium was replaced with 18âml fresh medium and the cells were transfected using the PEI (1âmgâmlâ1, Polysciences, 23966-2) transfection method. The individual gene library (25.5âµg) was transfected together with the lentiviral packaging plasmids psPAX2 (32.12âµg) and pCMV-VSV-G (9.44âµg) per 15âcm plate. psPAX2 was a gift from D.âTrono (Addgene, plasmid 12260; http://n2t.net/addgene:12260; RRID:Addgene_12260). pCMV-VSV-G was a gift from B.âWeinberg (Addgene, plasmid 8454; http://n2t.net/addgene:8454; RRID:Addgene_8454). One day (20â24âh) after transfection, the medium was changed to NeurobasalâA with penicillinâstreptomycinâglutamine. After another 20â24âh, lentivirus containing supernatant was collected and stored at 4â°C and fresh medium was added to the 293T cells for another collection after 24âh. Both supernatants were then combined, filtered through a 0.45âµm polyvinylidene fluoride filter (Millipore Sigma, SE1M003M00) and frozen at â80â°C in 5âml aliquots. For lentiviral transduction, young qNSCs were plated onto 6-well PDL pre-coated plates at a density of 1.75âÃâ106 cells per well (for control lentivirus), 10âcm PDL pre-coated plates at the density of 1.0âÃâ107 cells per plate (for Slc2a4-targeting lentivirus) or 12-well PDL pre-coated plates at a density of 4.0âÃâ105 cells (for Npb and B3galnt2 targeting lentivirus). NSCs were kept in qNSC medium for 4âdays (with medium changes every other day) before transduction. After removing medium, viral supernatants (2âml for 6-well plates, 10âml for 10âcm plates and 1âml for 12-well plates) were thawed at room temperature and mixed with 8% of B27 minus vitamin A, bFGF (80ângâmlâ1) and BMP4 (200ângâmlâ1). qNSCs were incubated with lentiviral medium for 24âh. After removing lentiviral medium after 24âh, a second lentiviral transduction was repeated the next day. After two consecutive transductions, qNSCs were washed once with NeurobasalâA medium and then cells were kept in qNSC medium for 7âdays to allow recovery and CRISPR editing. To select for a population of cells that was infected by the lentivirus, 1.0âμgâmlâ1 of puromycin (Sigma-Aldrich, P8833) was added to the cultures for 3âdays, with medium changes every day. To assess knockout efficiency, we isolated genomic DNA as described above for experimentâ1. We amplified genomic DNA with primer pairs roughly 150â250âbp upstream and 300â450âbp downstream of sgRNA editing site (see source data for list of primers) using GoTaq Green master mix (Promega, M7123) and the following amplification program: 30âs of annealing step at 55â°C and 1âmin of extending step at 72â°C for 40 cycles total.
In both experimentâ1 and experimentâ2, PCR amplicons were Sanger sequenced using the respective forward primers (source data). We then analysed knockout efficiency using the DECODR (v.3.0) online tool (https://decodr.org/)93. Each sgRNA was analysed separately, and the editing efficiency is indicated in source data. Individual sgRNAs that had an editing efficiency with a r2 value less than 0.6 from DECODR (v.3.0) are indicated as low confidence (LC) in source data and marked with a hash symbol in Extended Data Fig. 1n. Individual sgRNAs that were not detected by DECODR (v.3.0) in the Sanger sequencing trace are indicated as not detected (ND) in source data and not included as data points in Extended Data Fig. 1n. Finally, we note that the percentage of knockout per gene is probably also underestimated due to the fact that larger indels that span sgRNA cutting sites are not taken into account by DECODR.
In vivo gene knockout experiments
Stereotaxic surgeries were performed to inject virus into the lateral ventricle of mice. For these experiments, old Cas9 mice were used, except for one experiment for which old wild-type mice were used (see the section âLaboratory animalsâ). Surgeries were performed on heating pads with isoflurane-induced anaesthesia, with a Kopf (Model 940) stereotaxic frame, World Precision Instruments (UMP3T-1) UltraMicroPump3, Hamilton 1710RN 100âμl syringe with 30âg Small Hub RN needle with a point 2 bevelled end. Injections were made at the following coordinates, relative to bregma: lateral 1âmm, anterior 0.3âmm and ventral depth 3âmm from the skull surface. After drilling the skull and inserting the needle into position, we waited 5âmin before injecting the virus. We injected 3âµl of equal titre virus at a rate of 10ânlâsâ1. We waited 7âmin after injection before removing the needle and suturing the skin. Animals were administered a single dose of buprenorphine SR (0.5âmgâkgâ1) for postoperative pain management and monitored for 1âweek after surgery until full recovery. For labelling of proliferating NSC progeny, we injected animals intraperitoneally weekly with EdU (Thermo Fisher Scientific, A10044, 50âmgâkgâ1, dissolved in sterile PBS), starting 1âweek after surgery. We used both male and female mice for in vivo testing, always making a note of the sex for each experiment. We did not observe major differences in results between sexes, and plots include data from both sexes.
Influence of the anaesthetic
In our pilot experiments, we performed some surgeries with ketamineâxylazine anaesthesia instead of isoflurane for the relative ease of use, which we consider resulted in marked impairment of neurogenesis in both young and old animals when assessed in downstream screen analyses. In brief, we performed our in vivo screening as outlined above, but we could detect only very few sgRNAs in the olfactory bulb 5âweeks after injection when the mice had been anaesthetized with ketamineâxylazine. We interpreted the lack of sgRNA detection in the olfactory bulb as an indication that not many NSCs were able to activate and migrate to the olfactory bulb in those conditions. We repeated the experiments with ketamineâxylazine 2 times, in around 20 animals, always observing an impairment in sgRNA detection in the olfactory bulb after 5âweeks. When the same virus was injected into same age and background mice under isoflurane anaesthesia, we detected a greater diversity and abundance of sgRNAs in the olfactory bulb 5âweeks later. We therefore did not perform any surgeries presented in this article with ketamineâxylazine anaesthesia, but used isoflurane instead.
At the end point of in vivo experiments, mice were either killed for sequencing of sgRNAs in the brain (in vivo subscreens) or for immunofluorescence imaging (see the section âIn vivo immunofluorescence experimentsâ) of the olfactory bulb and other brain regions (single gene knockout experiments). For sequencing sgRNAs in the brain, mice were killed either 1â2âdays after injection or 5âweeks after injection and their brains were immediately removed and subdissected for genomic DNA extraction. We used a scalpel to cut off the olfactory bulbs and to cut an approximately 1âmm thin slice of the outer cortex as well as the outer cerebellum. We then subdissected out the SVZ niche. We took each tissue and minced it with around 100 cuts of a scalpel and proceeded to extract genomic DNA according to the manufacturerâs protocol (Qiagen QIAamp DNA micro kit, 56304). The genomic DNA was then processed for sgRNA amplification and sequencing as outline in the section âsgRNA PCR amplification and sequencingâ.
Immunofluorescence staining of brain sections, image analysis and quantification
Brain sections in the olfactory bulb and SVZ
For immunofluorescence straining of brain sections, young and old anaesthetized mice were first subjected to intracardiac perfusion with 4âml of heparin (Sigma Aldrich, H3149-50KU) and then 25âml 4% paraformaldehyde (PFA) (Electron Microscopy Science, 15714) in PBS. Brains were then removed and further fixed for 16âh by submerging in 4% PFA at 4â°C. Brains were then washed 3 times in PBS and placed in a conical tube with a 30% sucrose (Sigma-Aldrich, S3929-1KG) in PBS solution for 2â3âdays until sinking to bottom of conical tube. The brains were then embedded in optimal cutting temperature (OCT) compound (Electron Microscopy Sciences, 62550-12) for cryosectioning. Brain coronal sections were taken at 20âµm thickness (Leica, CM3050S). For assessing neurogenesis in the olfactory bulb, every tenth section was used. Thus, imaging was performed every 200âµm across the entire olfactory bulb. For assessing different cell types in the SVZ, we began taking sections at the most anterior part of the lateral ventricle, and every tenth section was used. Thus, imaging was performed every 200âµm across the SVZ.
Immunofluorescence staining of brain sections
For immunofluorescence staining, sections were brought to room temperature and then washed once with PBS and then permeabilized with ice-cold methanol and 0.1% Triton X-100 (Fisher Scientific, BP151) for 15âmin. All samples were stained at the same time. Slides were washed 3 times with PBS and then treated with ClickIt reagents (for EdU) or put straight into antibody blocking solution. For Click-It EdU staining (Thermo Fisher Scientific, C10337/C10639/C10634), we placed 50â70âµl of reaction cocktail from this kit onto the tissue and incubated in humidified chamber at room temperature for 30âmin. Slides were then washed 3 times in PBS before blocking for antibodies. Slides were treated with 50â70âµl blocking solution (5% normal donkey serum (NDS, ImmunoReagents, SP-072-VX10), 1% BSA (Sigma-Aldrich, A1595-50ML), 8.5âml PBS) in a humidified chamber at room temperature for 30âmin. Blocking solution was replaced with antibody solution consisting of blocking solution with antibodies as follows: mCherry (Invitrogen, M11217, clone 16D7) 1:500, GFAP (Abcam, 53554) 1:500, GFP (Abcam, 13970) 1:500, GLUT4 (for in vivo staining R&D Systems, MAB1262, clone 1F8) 1:500, Ki67 (Invitrogen, 14-5698-082, clone SolA15) 1:500, STX4A (Santa Cruz Biotechnology, sc-101301, clone QQ-17) 1:500, GFP (Abcam, 13970) 1:500, mouse IgG (Santa Cruz SC-3877, lot: L1916) 1:500, NeuN (Millipore, MAB377 clone A60) 1:500, S100a6 (Abcam, ab181975, clone EPR13084-69) 1:500, Tuj1 (BioLegend, 802001) 1:500, Olig2 (R&D Systems, AF2418) 1:100, Sox10 (Abcam, Ab180862, clone EPR4007-104) 1:100, calretinin (Abcam, Ab244299) 1:500, Dcx (Cell Signaling Technology, 4604) 1:500. We tested two mCherry antibodies (Abcam, ab213511 clone EPR20579; Invitrogen, M11217 clone 16D7) and we found that Invitrogen, M11217 was better for immunostaining for brain sections. After primary staining in dark for 2âh in humidified chamber at room temperature or 16âh at 4â°C, slides were washed 3 times in PBS before staining with secondary antibodies. Secondary antibodies were diluted in blocking solution and consisted of Alexa 488/594/647 conjugated antibodies (Fisher Scientific, A21202, A21206, A21209, A21447, A31571, A31573) 1:500, and DAPI (1âmgâmlâ1, Fisher Scientific 62248) 1:5,000. We added 50â70âµl of secondary antibody mix to cover the section and incubated in the dark for 2âh in a humidified chamber at room temperature or 16âh at 4â°C. Slides were then washed 3 times with PBS 0.2% Tween for 10âmin, washed 3 times with PBS for 5âmin and then mounted using ProLong Gold (20â40âµl, Thermo Fisher Scientific, P36931), dried for 2âh and sealed with nail polish. To allow for quantification of the immunofluorescence staining, we paid special attention to stain all the brain sections from different groups (for example, young and old) in the same way and at the same time.
Confocal imaging of immunofluorescence staining in brain sections
Images were captured using a Zeiss LSM 900 confocal microscope with a Ã10, Ã20 or Ã63 objective, with Zen blue edition (v.3.0). The exposure and gain settings for each channel and antibody were set at the beginning of each imaging session and remained the same for all animals and treatments. We randomized the order in which we imaged the slides and we ensured that different treatments and age groups were all imaged in the same session on the same day. The imaging was not performed in a blinded manner. We did not select areas to image. We imaged and quantified serial sections. Confocal imaging was done every 200âμm across the entire olfactory bulb or SVZ region.
Image analysis and quantification of immunofluorescence staining in brain sections
For image analysis, we used the open-source software QuPath (https://qupath.github.io/)94. This approach allowed us to set the thresholds and quantification parameters on training images and then ran the same analysis across all sections, samples and treatments in an automated manner. Many cells (>100 cells per section in vivo) and many sections (>50 sections per age group in vivo) were counted in an unbiased manner using an automated pipeline (in QuPath).
Quantification of GLUT4 depletion efficiency in the SVZ niche by immunostaining in vivo
For quantifying GLUT4 depletion efficiency in the SVZ niche, we first annotated a polygonal line around the SVZ NSC niche, creating an analysis region about 5â20âcells deep from the ventricle wall. We then performed the âanalyseâcell detectionâ function, detecting cells in the image based on DAPI staining, using the program default settings, expanding the cell nuclei 5âµm in the âcell parametersâ section. We then trained two independent object classifications for GFAP+ and mCherry+ cells, adjusting the thresholds to detect positive cells that were apparent by eye. We combined the GFAP+ and mCherry+ objects into a single composite classifier and ran it on all annotated images and treatments. The results were output as annotation detections. The annotation detections were used to display the GLUT4 channel cell mean fluorescence intensity for GFAP+mCherry+ compared with GFAP+mCherryâ populations in the different treatments.
Quantification of newborn neurons in the olfactory bulb by immunostaining in vivo
For quantification of newborn neurons in the olfactory bulb, we first annotated a polygon line immediately beneath the olfactory bulb mitral cell layer to focus the analysis within the inner layers of the olfactory bulb, where newborn neurons arrive. We then performed the âanalyseâcell detectionâ function, detecting cells in the image based on DAPI staining, using the program default settings, expanding the cell nuclei 5âµm in the âcell parametersâ section. We then trained three independent object classifications for mCherry+, EdU+ and NeuN+ cells, adjusting the thresholds to detect positive cells that were apparent by eye. We combined the mCherry, EdU and NeuN objects into a single composite classifier and ran it on all annotated images and treatments. The results were output as annotation measurements and annotation detections. The annotation measurements were used for graphs depicting the number of NeuN+mCherry+EdU+/total EdU+ cell numbers for each treatment, and the annotation detections were used to display the NeuN channel cell mean fluorescence intensity for EdU+mCherry+ populations in the different treatments.
Quantification of different cell numbers in the SVZ niche by immunostaining in vivo
For quantifying different cell numbers in the SVZ niche with Slc2a4 sgRNA treatment compared with control, we first annotated a polygonal line around the SVZ NSC niche, creating an analysis region about 5â20 cells deep from the ventricle wall. We then performed the âanalyseâcell detectionâ function, detecting cells in the image based on DAPI staining, using the program default settings, expanding the cell nuclei 5âµm in the âcell parametersâ section. We then trained three independent object classifications for GFAP+, Ki67+ and S100a6+ cells, adjusting the thresholds to detect positive cells that were apparent by eye. We combined the GFAP+, Ki67+ and s100a6+ objects into a single composite classifier and ran it on all annotated images and treatments. The results were output as annotation measurements. The annotation measurements were used for graphs depicting the sgRNA treatment and impact on number of each cell type: qNSCs (GFAP+S100a6+Ki67â), aNSCs (GFAP+S100a6+Ki67+), neuroblasts (GFAPâKi67+) and astrocytes (GFAP+S100a6â) for each condition.
Quantification of GLUT4 protein levels in different cell types of the SVZ niche by immunostaining in vivo
For quantification of GLUT4 fluorescence intensity in different cell types of young and old mice in vivo, we first annotated a polygonal line around the SVZ NSC niche, creating an analysis region about 5â20 cells deep from the ventricle wall. We then performed the âanalyseâcell detectionâ function, detecting cells in the image based on DAPI staining, using the program default settings, expanding the cell nuclei 5âµm in the âcell parametersâ section. We then trained two independent object classifications for Ki67+ (or S100a6+, for NSC specific labelling experiments) cells and GFAP+ cells, adjusting the thresholds to detect positive cells that were apparent by eye. We combined the Ki67 (or S100a6) and GFAP objects into a single composite classifier and ran it on all annotated images and treatments. The results were output as annotation detections. The annotation detections were used to display the GLUT4 channel cell mean fluorescence intensity for GFAP+Ki67+ (aNSCs), GFAP+Ki67â (qNSCs/astrocytes), GFAP–Ki67+ (neuroblasts), GFAPâKi67â (other cells, including ependymal and microglia) or GFAP+S100a6+ (NSCs) populations across different aged mice. The GLUT4 antibody we used for immunostaining of brain sections (R&D Systems, MAB1262, clone 1F8) was validated in vivo by the Slc2a4 knockout (see above).
For all experiments, the output numbers displayed on the graphs were derived from the average of all serial section images across a biological replicate (one mouse), biological sample values were then analysed for significance by two-tailed MannâWhitney test.
Immunofluorescence staining of primary cell cultures of NSCs and quantification
Immunofluorescence staining of NSC cultures
For immunofluorescence staining of primary cell cultures of NSCs, we seeded 2.5âÃâ105 aNSCs or 2âÃâ105 qNSCs onto PDL (50ângâmlâ1, Sigma-Aldrich, P6407) pre-treated (30âmin, followed by 3à PBS wash) coverslips in each well of a 24-well plate. The qNSCs were plated 7âdays before fixation, the aNSCs were plated 24âh before fixation. For fixation, cells were washed once with PBS and then 500âμl of 4% PFA (Electron Microscopy Science, 15714) was added for 30âmin of incubation at room temperature. Cells were washed 3 times with PBS and then permeabilized with 0.1% Triton X-100 (Fisher Scientific, BP151) in PBS for 15âmin shaking at room temperature. Coverslips were washed twice with PBS and then processed for antibody staining. Coverslips were placed on a 45âµl drop of primary antibody solution consisting of 1% BSA in PBS with primary antibodies as follows: GLUT4 (Abcam, 33780) 1:500, Ki67 (Invitrogen, 14-5698-082) 1:500, STX4A (Santa Cruz Biotechnology, QQ-17) 1:500. After 1âh of incubation in the dark at room temperature, slides were washed 3 times in PBS shaking for 5âmin at room temperature. Slides were then placed on 45âµl drop of secondary antibodies in 1% BSA in PBS consisting of Alexa 488/594/647 conjugated antibodies (Fisher Scientific, A21206, A21209, A31571) 1:500, and DAPI (1âmgâmlâ1, Fisher Scientific 62248) 1:5,000. After 1âh of incubation at room temperature in the dark, slides were washed 3 times with PBS before mounting with ProLong Gold, dried for 2âh and sealed with nail polish. To allow quantification of immunofluorescence staining, we paid special attention to stain all coverslips in the same way and at the same time.
Confocal imaging of immunofluorescence staining in NSC cultures
Images were captured using a Zeiss LSM 900 confocal microscope with a Ã10, Ã20 or Ã63X objective. The exposure and gain settings for each channel and antibody were set at the beginning of each imaging session and remained the same for all samples and treatments. We randomized the order in which we imaged the slides, and we ensured that different treatments and age groups were all imaged in the same session on the same day. The imaging was not done in a blinded manner. We did not select areas to image. We randomly selected ten areas of each coverslip to image. For image analysis, see the section âImmunofluorescence image analysisâ.
Image analysis and quantification of immunofluorescence staining in NSC cultures
For image analysis, we used the open-source software QuPath (v.0.2.3) (https://qupath.github.io/)94. This approach allowed us to set the thresholds and quantification parameters on training images and then ran the same analysis across all sections, samples and treatments in an automated manner. For the in vitro GLUT4 and STX4A quantifications, we selected the entire image as the analysis annotation. We then performed the âanalyseâcell detectionâ function, detecting cells in the image based on DAPI staining, using the program default settings, expanding the cell nuclei 5âµm in the âcell parametersâ section. The results were output as annotation detections. The annotation detections were used to display the GLUT4 and STX4A cell mean fluorescent intensity for each proteinâs channel in each cell culture type and age group. For all experiments, the output numbers from different images were averaged across a biological replicate (one NSC culture), biological sample values were then analysed for significance by two-tailed MannâWhitney test. We note that the age-dependent increase in GLUT4 and STX4A proteins was not large in qNSC cultures. In addition, we were not able to detect significant changes in Slc2a4 by RTâqPCR and western blotting in young and old qNSCs. This lack of detection is probably due to sensitivity issues: single-cell RNA sequencing and immunofluorescence staining are single-cell-based assays, which can be more sensitive than bulk assays (such as RTâqPCR and western blotting) in capturing small differences in transcript or protein expression.
Expression of glucose transporter genes and fatty acid oxidation gene signature in single-cell RNA sequencing
To test the expression of Slc2a4 and other glucose transporter genes, we retrieved the raw counts from our most recent single-cell RNA sequencing dataset from the SVZ neurogenic niche from young and old mice70 and used a subset of the data containing only the control (sedentary) animals across young and old ages (i.e. O_Control and Y_Control in the AgeCond metadata column). We normalized the counts data by dividing each cell by its total expression, scaling up to 105 total counts per cell, and then taking the log-transform of the normalized counts with an added pseudocount. For comparisons across cell types, we used the pre-existing cell-type annotations: Astrocyte_qNSC, aNSC_NPC and Neuroblast70. For comparisons across age, we compared old (O_Control) and young (Y_Control) animals using the pre-existing age annotations in the dataset70. For statistical comparisons of the mean expression across conditions, we used the two-sample Welchâs t-test from stat_compare_means(). Welchâs t-test is designed for unequal population variances. To compute the log fold change, we divided the average expression in the old cells by the average expression in young cells and then took the log2-transform of the resulting ratio. To compute the fatty acid oxidation gene signature, we summed the expression of the 19 genes from the fatty acid oxidation signature published in ref. 95 for each cell in the published single-cell RNA sequencing dataset from SVZ neurogenic niches of young and old mice70. We then compared the fatty acid oxidation gene signature levels across old and young qNSCs/astrocytes using the two-sample Welchâs t-test, which is designed for unequal population variances.
Expression of Slc2a4 RNA in bulk RNA sequencing (in vitro)
To test the expression of Slc2a4 in vitro, we retrieved the RNA sequencing normalized counts from in vitro qNSCs and aNSCs from a published dataset51. To calculate significance between qNSCs and aNSCs, we used a two-sided MannâWhitney test.
Glucose uptake assays
For qNSCs, we seeded 40,000 cells per well and for aNSCs we seeded 10,000 cells per well (aNSCs do not stick to the plate as well and will double every 16â24âh, so we seeded fewer cells to achieve similar density to qNSCs at the time of analysis) on PDL (Sigma-Aldrich, P6407) pre-coated 96-well plates, performing the assay 3âdays after seeding. Duplicate wells were seeded and used for cell count normalization at time of glucose uptake assay. For knockout experiments, 1âÃâ105 qNSCs were plated per well on PDL pre-coated 96-well plates in qNSC medium 6âdays before infection with lentivirus to express sgRNA, for which 1âμl of concentrated virus was added to the culture medium for 16âh to achieve about 100% infection of the cells. We then assessed glucose uptake either 4âdays or 8âdays after infection using two different types of assays (see below).
Colorimetric glucose uptake assay
For the colorimetric glucose uptake assay (Glucose Uptake-Glo Assay, Promega, J1342; Fig. 4f,h), experiments were performed according to the manufacturerâs protocol, with the following details. Cells were pre-treated for 1âh of qNSC/aNSC culture medium without glucose. Culture medium was then replaced with 50âµl of qNSC/aNSC medium containing 1âmM 2-DG (provided in the Glucose Uptake-Glo kit from Promega (J1342)) reagent for 10âmin in an incubator (humidified, 37â°C, 5% CO2). The 2-DG medium was then removed and 50âµl of PBS was added before carrying out the remainder of the assay according to the manufacturerâs protocol. All media treatments and reagent exchanges were pre-aliquoted into an empty 96-well plate, such that we could add the treatment to entire rows of cells at once using a multi-channel pipette to ensure that the duration of treatment was equivalent across different cell types and ages. The luminescence of the cells was measured with 0.5âs readings using a Varioskan LUX multimode plate reader. Owing to different treatments having effects on cell numbers, plate readings in some cases (mentioned in figure legends) required normalization to the cell counts (Countess II cell counter, Thermo Fisher Scientific) based on duplicate wells. We performed glucose uptake experiments on different numbers of NSCs and observed a linear correlation between relative light units and cells plated.
Fluorescent glucose uptake assay
For the fluorescent 2-NBDG glucose uptake assay (fluorescent 2-NBDG [2-(N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)Amino)-2-deoxyglucose] (Fisher, N13195; Extended Data Fig. 6f), cells were placed in glucose-free medium for 1âh and then treated with 200âµM 2-NBDG for 30âmin at 37â°C, and then analysed by flow cytometry at excitation/emission maxima of around 465/540ânm, with DAPI added in the medium to eliminate dead cells.
Assessing GLUT4 depletion efficiency at the protein level in vitro
Western blot to assess GLUT4 depletion efficiency at the protein level
Young aNSCs were seeded onto PDL-coated 10âcm plates at a density of 1âÃâ107 cells per plate and transferred into qNSC medium. Medium was changed every 2âdays. After 4 or 5âdays in qNSC medium, qNSC cultures were infected with lentivirus expressing control sgRNAs or Slc2a4 sgRNAs (5 sgRNAs). Seven days after infection, 1.0âμgâmlâ1 puromycin (Sigma-Aldrich, P8833) was added to the cultures for 3âdays, with medium changes every day, to select for infected cells. Then, the cells were washed with PBS and incubated on ice with ice-cold 1à lysis buffer (50âmM Tris-HCl, pHâ8, 150âmM NaCl, 0.5% sodium deoxycholate and 0.5% Triton-X 100) and 1à protease inhibitor (Thermo Fisher Scientific, 87786) for 10âmin and cells were scraped off of the plate. Lysates were centrifuged at 10,000g for 10âmin at 4â°C. The supernatant was removed and preserved, then the protein concentration was quantified using a BCA assay (Thermo Fisher Scientific 23225). To load the samples, 4à LDS buffer (Invitrogen NP007) with 1âmM DTT (Sigma-Aldrich 10197777001) was added to lysates with equal concentrations of protein and the mix was incubated at 95â°C for 7âmin; 25âμg of protein was added to each lane. Proteins were separated by SDSâPAGE in MOPS buffer (Invitrogen NP0001) on precast 4â12% Bis-Tris polyacrylamide gels (InvitrogenNP0323BOX). Proteins were transferred onto nitrocellulose membranes. Membranes were incubated for 30âmin at room temperature in blocking buffer (PBSâ+â3% w/v non-fat dry milkâ+â0.2% Tween-20). Given that GLUT4 and the loading control (β-actin) have a similar molecular weight, we performed western blotting in a sequential manner (first GLUT4 and then β-actin). Primary antibodies to GLUT4 (1:500, Invitrogen PA1-1065) were diluted in blocking buffer and incubated overnight at 4â°C. After three washes in PBSâ+â0.2% Tween-20, goat anti-rabbit 800CW (1:10,000, Li-Cor 925-32211) in blocking buffer were incubated for 1âh at room temperature and washed 3 times in PBSâ+â0.2% Tween-20. Detection was performed on a Li-Cor Odyssey FC imaging system with the 800 channel for 10âmin. Then primary antibodies to β-actin (1:40,000, Abcam ab6276) as a loading control were added for 1âh at room temperature and washed 3 times in PBSâ+â0.2% Tween-20. Goat anti-mouse 680CW (1:10,000, Li-Cor 925-68070) in blocking buffer was incubated for 1âh at room temperature and washed 3 times in PBSâ+â0.2% Tween-20. Detection was performed on a Li-Cor Odyssey FC imaging system with the 700 channel for 30âs. We used ImageJ to quantify the intensity of the GLUT4 and β-actin bands, and the intensity of the GLUT4 band was divided by the intensity of the corresponding β-actin band for each sample. We note that we used different GLUT4 antibodies used for immunofluorescence and western blot experiments because the GLUT4 antibodies used for immunofluorescence did not work for western blotting. This is most likely due to the fact that proteins are in their native form in immunofluorescence experiments but are denatured in western blot experiments. Both GLUT4 antibodies for immunofluorescence and western blotting were both validated using Scl2a4 (GLUT4 knockout) (Fig. 3aâd and Extended Data Fig. 6j).
FACS to assess GLUT4 depletion efficiency at the protein level
We plated NSCs on PDL-coated 24-well plates at the density of 3âÃâ105 cells per well and added qNSC medium for 4âdays. After 4âdays in quiescence, lentivirus expressing control sgRNAs or sgRNAs to Slc2a4 (5 sgRNAs) was added to qNSCs for overnight infection, and the cells were kept in qNSC medium for another 6âdays. After 6âdays (to leave time for infection and knockout to occur), the cells were dissociated with Accutase and placed in 500âμl of medium in a 24-well format. FACS was performed by mixing the primary GLUT4 antibody (R&D Systems, MAB1262) at a 5:1 ratio with secondary anti-IgG AlexaFluor647 for 10âmin on ice, in the dark. The antibody mix was added to live cells in culture at a dilution factor of 200à (502.5âμl total volume) and incubated in a cell culture incubator (37â°C, 5% CO2) for 30âmin. The cells were then fixed by adding 500âμl of PBSâ+â1% PFA to each well, without shaking the cells. Cells were incubated at room temperature for 20âmin in the dark, and then analysed by FACS (BD, LSRFortessa). FACS quantification was done by gating first on mCherry+ cells (infected cells).
ECAR and OCR
To measure ECAR and OCR, we seeded 80,000 NSCs into qNSC medium in a PDL pre-treated well of a 96-well plate. The cells were maintained in quiescence for 4âdays with medium exchanges at 24 and 72âh after seeding. The cells were then treated with unconcentrated equal titre lentivirus (with control sgRNAs or sgRNAs to Scl2a4) for 16âh of overnight infection. The cells were placed back into qNSC medium for 48âh before running the metabolic assay. For ECAR, assays were run according to manufacturerâs protocol (Glycolysis assay, Abcam, Ab197244), with the following parameters. The cells were placed in a CO2-free incubator at 37â°C for 3âh before running the assay. Fluorescence was measured using a Tecan Spark plate reader with the following settings: instrument was pre-warmed to 37â°C 1âh before the run, the run mode parameters were as follows: kinetic, kinetic duration 90âmin, interval time 1âmin and 30âs, excitation wavelength 380, excitation bandwidth 20, emission wavelength 615, emission bandwidth 10. The slope of the fluorescence detected over the period of linear increase was calculated for each sample. For OCR, assays were run according to the manufacturerâs protocol (Extracellular Oxygen Consumption Assay, Abcam, Ab197243) with the following parameters. The extracellular O2 consumption reagent was used at 1/15 dilution (10âµl added to 150âµl of sample in a 96-well plate). High-sensitivity mineral oil was pre-warmed to 37â°C 30âmin before use, and 2 drops were used for each well before running the assay on plate reader. Fluorescence was measured using a Tecan spark plate reader with following measurements: excitation 380â±â20ânm, emission 650â±ââ20ânm, kinetic duration 1âh and 30âmin, interval time 1âmin and 30âs. The slope of the fluorescence detected over the period of linear increase was calculated for each sample.
Transient glucose starvation
NSCs were placed in qNSC medium for 4âdays, exposed to lentivirus to express sgRNAs targeting Slc2a4 or unannotated genomic regions (control). Then 6âdays after infection, the cell medium was replaced with standard complete qNSC medium with glucose or modified to have no glucose (NeurobasalâA medium, Thermo Scientific, A2477501, no d-glucose, no sodium pyruvate, supplemented with 1Ã sodium pyruvate, Fisher Scientific, 11-360-070) for 48âh, at which point the medium was replaced with standard complete aNSC medium (with normal glucose concentration (4,500âmgâlâ1) in NeurobasalâA medium, Thermo Fisher 10888-022) and the cells were allowed to activate for 4âdays before intracellular FACS analysis with Ki67.
Effect of 2-DG on young and old NSC activation
To test the effect of 2-DG on young and old NSC activation, we performed qNSC activation experiments in a 24-well plate format. Primary cultures of NSCs were derived from a pool of 2 young (3â4âmonths old) or old (18â21âmonths old) mice (1:1 mix of male and female). We seeded 2âÃâ105 NSCs in each well of a 24-well plate. After 4âdays in qNSC medium (with qNSC medium changes every 2âdays), 2-DG (2âmM final concentration, Sigma, D8375) was added to the medium for 36âh of treatment, with one exchange at the 24-h time point. After 36âh of treatment, the cells were washed 1à in PBS and then transitioned to aNSC medium for activation. aNSC medium was exchanged once after 48âh and then Ki67 intracellular FACS was performed at dayâ4 after treatment to assess NSC activation efficiency as described above. Pâvalues were determined by two-tailed MannâWhitney test.
Statistical analyses
We did not perform randomization, but for all experiments, young and old conditions were processed in an alternate manner rather than in two large groups to minimize the group effect. We did not perform power analyses, although we did take into account previous experiments to determine the number of animals needed. To calculate significance for experiments, all tests were two-sided MannâWhitney tests, unless otherwise indicated. Results from individual experiments and all statistical analyses are included in the source data.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.