Cell culture
The hTERT RPE-1, HEK293(T) and HeLa S3 cell lines were cultured in Dulbecco’s modified Eagle’s medium/F12 (Merck) supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific) and 100 U ml−1 of streptomycin and 100 mg ml−1 of penicillin (Gibco). HeLa S3 dCas9–ZIM3 cells were cultured in the presence of 200 µg ml−1 of hygromycin to maintain selection. K562 dCas9–KRAB cells were cultured in RPMI-1640 with 25 mM HEPES and 2.0 g l−1 of NaHCO3 in 10% FBS, 2 mM glutamine, 100 U ml−1 of streptomycin and 100 mg ml−1 of penicillin. All cell lines were grown under 3% oxygen. Cells were routinely tested for mycoplasma.
Cell line generation
The hTERT RPE-1 TP53 KO cells were obtained from the Stephen Jackson lab (Cambridge). RPE-1 TP53 KO dCas9–KRAB cells were generated by transduction with a lentiviral vector encoding the dCas9–KRAB and a blasticidin resistance cassette with a low multiplicity of infection (MOI). Cells were selected using blasticidin, and single-cell clones were seeded by cell sorting. The resulting clones were validated for CRISPRi activity by CD55 knockdown efficiency. RPE-1 TP53 KO, FANCM KO and SMARCAL1 KO cell lines were generated using CRISPR–Cas9. SgRNAs were selected from the Vienna BioCenter (VBC) website and were in vitro transcribed with T7 RNA polymerase64. Editing of cells was done, as described previously65. A Lonza Amaxa nucleofector was used with P3 solution and program EA104. Clones were single-cell seeded by serial dilution, and clones were selected using Sanger Sequencing and ICE analysis (Synthego). KO of FANCM and SMARCAL1 was validated by next-generation sequencing and western blotting. A pool of HeLa S3 dCas9–ZIM3-expressing cells was generated by transduction with a lentiviral vector encoding dCas9–ZIM3 and a hygromycin resistance cassette, followed by selection with 200 µg ml−1 of hygromycin.
Cloning and mutagenesis
For RNase H1 wild-type or WKKD mutant (W43A, K59A and K60A), cDNAs from Addgene plasmid nos. 111906 and 111905 were polymerase chain reaction (PCR) amplified. The resulting fragments and mCherry cDNA amplified from Addgene plasmid no. 102245 were assembled using Gibson assembly. Empty and FANCM (wild type and K117R) pLVX-TetOne-Puro plasmids were a generous gift from the Claus Maria Azzalin Lab66. pAcGFP-C1–RPA123–P2A was a generous gift from the Jiri Lukas laboratory. Point mutants were created by site-directed mutagenesis. Internal deletion mutants were generated by inverse PCR, followed by T4 polynucleotide kinase phosphorylation and blunt-end ligation. 3×Flag–SMARCAL1, PCNA, FEN1 and LIG1 cDNAs were cloned into the lentiviral vector pLVX-TetOne-Puro (Clontech) using Gibson assembly. For all Gibson assemblies, the New England Biolabs (NEB) HiFi Assembly Master Mix was used. All PCR reactions were performed using Q5 DNA Polymerase (NEB).
Guide RNA cloning
CRISPRi sgRNAs were cloned into either BFP-, GFP- (Addgene plasmid nos. 60955 and 111596) or mCherry-containing lentiviral vectors or, for dual-guide expression, into pJR103 (Addgene plasmid no. 187242). A neomycin resistance cassette-containing lentiviral vector was used for sgRNAs targeting RAD18 (Fig. 2c,d,f,g and Extended Data Figs. 5h,i, 6e–f and 7a,b,e–g), LIG1 (Fig. 2e and Extended Data Fig. 7d,h), and FANCM (Figs. 3a and 4d,e and Extended Data Fig. 12g). For dual guide expression vectors, homologies with the backbone and protospacers were added to the insert from pJR98 (Addgene plasmid no. 187239; containing the constant region CR3 for the first sgRNA and hU6 promoter for the second sgRNA) using PCR. PCR products were assembled with JR103 using Gibson assembly. Forward and reverse oligos for each sgRNA were annealed by preincubation at 37 °C for 30 min with T4 polynucleotide kinase (NEB), followed by incubation at 95 °C for 5 min and then ramp down to 25 °C at 5 °C min−1. Annealed sgRNAs were ligated into the corresponding vector that had been digested with BstXI and BlpI restriction enzymes using T4 ligase (NEB). All sgRNA protospacers are listed in Supplementary Table 6.
Design of double-sgRNA CRISPRi SPIDR library
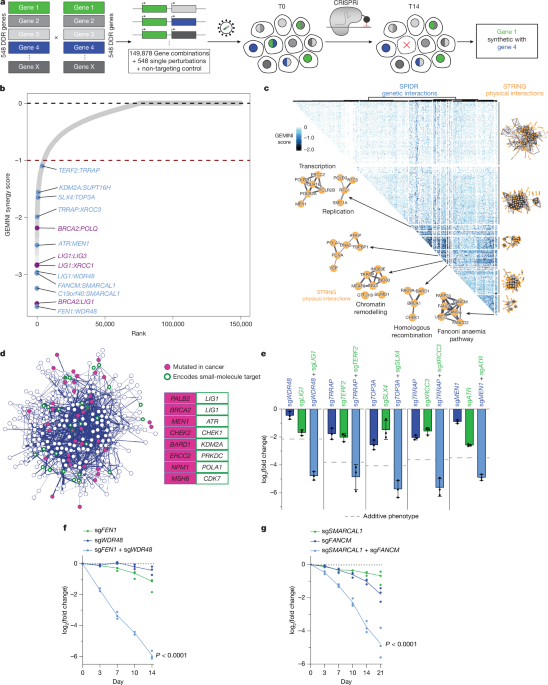
The genes to target in the SPIDR sgRNA library (n = 548) were selected by all genes with the gene ontology term ‘DNA repair’ (GO:0006281). For each gene, sgRNAs from the human CRISPRi-v2 library were ranked using a strategy described previously and a preliminary dataset of more than 50 CRISPRi screens10,12. Briefly, the genes were divided into three tiers. For genes essential for growth in K562 cells (tier 1), sgRNAs were ranked according to their growth phenotypes. For genes that had scored as significant hits in at least four CRISPRi screens (tier 2), sgRNAs were ranked according to their average phenotype across all screens in which the genes had scored as hits. For all other genes (tier 3), sgRNAs were ranked according to the regression scores from the human CRISPRi v.2.1 algorithm. For each gene, two sgRNAs were then selected for each transcription start site targeted in the human CRISPRi-v2 library, as follows.
For genes (n = 249) for which KO caused strong growth phenotypes in either RPE-1 cells or in at least five neuroblastoma cell lines, as assessed by querying data from 17 screens in neuroblastoma cell lines (identifiers NB5, NB6, NB7, NB10, NB13, NB17, NB69, CHP-212, SK-N-FI, SK-N-AS, SK-N-DZ, SK-N-SH, KP-N-YN, KP-N-YS, TGW, BE2-M17 and NH-12) in the Project Score database from the Cancer Dependency Map (https://score.depmap.sanger.ac.uk/, accessed August 2020), these two sgRNAs were (1) the top sgRNA identified using the algorithm above and (2) a mismatched variant of the top sgRNA, chosen to have an empirical relative activity of 0.47 ± 0.15 if an sgRNA existed for which the relative activity in this window had been measured or otherwise chosen as the mismatched variant with predicted activity closest to 0.5 out of all possible singly mismatched variants, with activity predictions derived from a convolutional neural network, as previously described11,67,68. Mismatched sgRNAs were required to have an on-target specificity score of at least 0.15, calculated as described previously11.
For genes (n = 299) for which KO did not cause strong growth phenotypes in both RPE-1 and more than 12 neuroblastoma cell lines, the top 2 sgRNAs from the algorithm above were selected.
From this set of sgRNAs, the double-sgRNA libraries were assembled in a programmed manner. Each unique combination of two genes was randomly assigned an orientation of ‘ab’ or ‘ba’. For each such a combination, all sgRNAs targeting the gene in position a were paired with all sgRNAs targeting the gene in position b. Each sgRNA targeting a given gene was also paired with all other sgRNAs targeting that gene. Each unique sgRNA in the library was furthermore paired with 15 different non-targeting negative control sgRNAs in both the ab and ba orientations. Finally, 225 pairs consisting of all possible combinations of 15 different non-targeting negative control sgRNAs were included as negative controls.
All sgRNA pairs were then ordered as an oligo pool from Agilent Technologies, containing the following constant sequences: AACTGCGATCGCTAATGTCCACCTTGTTG (upstream of sgRNA a), gtttcagagcgagacgtgcctgcaggatacgtctcagaaacatg (between sgRNA a and sgRNA b) and GTTTAAGAGCTAAGCTGGTTCTCCAGTGCCTTATT (downstream of sgRNA b).
Dual-guide library cloning
Dual-guide library cloning was performed, as described in a previous study12 with small modifications. After PCR amplification, oligos were BstXI/BlpI-digested and run on a polyacrylamide gel. The corresponding band was extracted, and DNA was purified before ligation into pJR103. The ligated DNA was precipitated using isopropanol (catalogue no. 17170576; Thermo Fisher Scientific) and transformed into MegaX Electrocomp Cells (catalogue no. C640003; Thermo Fisher Scientific). Bacteria were cultured in lysogeny broth overnight, and plasmid was collected with Plasmid Plus Midi Kit (QIAGEN). This intermediate library and pJR98 were digested using BsmBI, and the insert from pJR98 was ligated into the intermediate library to insert constant region CR3 for the first sgRNA and hU6 promoter for the second sgRNA. A library coverage of at least 30 times was continually maintained during the cloning process. The final library expresses the first sgRNA under the mU6 promoter with CR3, whereas the second sgRNA is expressed under a hU6 promoter with CR1.
Lentivirus packaging
Lentiviruses were produced in HEK293T cells. Briefly, HEK293T cells were transfected using polyethylenimine with the VSV-G envelope expressing plasmid (pMD2.G), packaging plasmid (psPAX2) and our transfer vector. Lentiviral supernatant was collected 48 and 72 h after transfection.
CRISPRi screens
The hTERT RPE-1 TP53 KO cells stably expressing dCas9–KRAB were transduced with the lentiviral library at an MOI of approximately 0.25 with a coverage of at least 350 cells per sgRNA in a medium supplemented with polybrene (10 μg ml−1). The next day, the culture medium was replaced with a puromycin-containing medium (15 μg ml−1) to select for transductants. Selection was carried out for 96 h, during which time cells were expanded and divided into two biological replicates. Hereafter, library coverage was maintained at approximately 500 cells per sgRNA combination. Four days after transduction, the background/time point 0 samples were collected and cell pellets were frozen (−80 °C). The remaining cells were seeded and continually subcultured when near 100% confluency for 14 days (approximately ten population doublings), at which time cell pellets were collected and frozen (−80 °C). Genomic DNA was isolated using the Gentra Puregene Cell Kit (QIAGEN). Integrated sgRNA regions were amplified by PCR using custom primers (Supplementary Table 7), and paired-end sequencing was performed on a NovaSeq 6000. Screens in HeLa S3 and K562 cells were performed, as described above, with the following modifications. Transduced HeLa S3 dCas–ZIM3 and K562 dCas9–KRAB cells were selected in a medium containing 1 or 5 μg ml−1 of puromycin, respectively, and selection was initiated 48 h after lentiviral transduction. HeLa S3 dCas9–ZIM3 cells were cultured in the presence of 200 µg ml−1 of hygromycin for the entirety of the screen.
Screen analysis
Raw FASTQ files were processed using seal (BBMap; B. Bushnell; sourceforge.net/projects/bbmap/) to search for sgRNA matches in the first 22 bp of each read with a hamming distance of 0 and a k-mer length of 20 bp. A custom Python script was used to parse the annotated FASTQ files and generate a count matrix containing all possible sgRNA pairs. The coverage per sgRNA pair was calculated to assess the quality of each sample (Extended Data Fig. 1a,b). Total counts were normalized using the median ratio method to estimate size factors using the non-targeting sgRNAs69. R v.4.1.2 was used to generate custom plots. Normalized sgRNA counts are provided in Supplementary Table 8.
Normalized counts were used to obtain log-fold changes (LFCs) from the GEMINI analysis. Count data were filtered with having a representation of 50 or more reads in the T0 sample of each replicate. LFCs were calculated using the GEMINI package with a pseudo-count of 10 and a modified gemini_calculate_lfc_mod function to prevent double normalization (see Supplementary Information). Essential sgRNAs were determined with having an average LFC of less than −3 at T14 when combined with non-targeting sgRNAs. Perfect and mismatched variants of these sgRNAs were included as separate genes in the analysis for Extended Data Fig. 2c to determine the recovery of genetic interactions for strong essential genes. Mismatched variants of strong essential sgRNAs showed a reduced signal at T14 and were able to recover more genetic interactions (Extended Data Fig. 2a–c). Therefore, count data were filtered for these candidates, and only the mismatched sgRNA variant was kept for all analysis. For HeLa S3 and K562, essential genes were defined with the same z-score cut-off as for RPE-1 LFC less than −3. Non-targeting sgRNAs were used as the negative control gene to build the GEMINI model. Sensitive and strong GEMINI scores were calculated with a scaling factor λ of 1 to identify synthetic lethal interactions of a broad range of genes. GEMINI scores greater than 0 were considered potential genetic interactions, and all combinations with a score of less than 0 were considered non-interacting and assigned a score of 0. The resulting GEMINI scores were given negative signs. For the genetic interaction heat map, the full SPIDR T14 dataset was filtered for genes having interactions with at least one other gene with a GEMINI sensitive score of 2 s.d. or more away from the mean non-zero score. Clusters were generated using agglomerative clustering with Ward variance minimization. Pairwise and higher physical interactions within each cluster were retrieved from STRING v.12 with a confidence cut-off of 0.4. UMAP embedding was calculated in R using the strong score across a symmetric version of the entire dataset, without prior dimension or variance reduction. A Venn diagram for the focused SPIDR screens was created using R with a GEMINI score cut-off of −1 or less for K562 cells and −0.5 or less for HeLa S3 cells.
Dual-colour flow cytometry assay
The indicated cells were transduced with sgRNA-containing lentiviruses at the same MOI. After 72 h, the cells were seeded in triplicate in six-well plates, and after an extra 24 h, the cell populations were analysed by flow cytometry (day 0) using an Attune NxT Flow Cytometer (Invitrogen). The cell populations were analysed at regular time intervals (as indicated in the figures) thereafter. For triple knockdown and cDNA experiments, cells were transduced with the relevant sgRNA-containing lentiviruses and selected with puromycin for 96 h before transduction with lentiviruses containing sgRNAs targeting synthetic lethal gene pairs. For cDNA experiments, expression was induced with 0.25–1 μg ml−1 of DOX 24 h before transduction with lentiviruses containing sgRNAs. The DOX-containing medium was refreshed every 48 h. Where drug treatments were performed, cells were treated 24 h after seeding on six-well plates (on day 0). All values were normalized to the corresponding untransduced control cells.
Clonogenic survival assays
The hTERT RPE-1 TP53 KO cells were co-transduced with lentiviruses containing sgRNAs that were co-expressed with either a puromycin or neomycin resistance cassette. After selection with puromycin (10 μg ml−1) and G418 (1.25 mg ml−1), 1,000 cells of each sample were seeded in triplicate on six-well plates. Where indicated, drug treatments were performed 24 h after seeding. Ten days later, cells were rinsed in PBS and stained with 0.5% (w/v) crystal violet in 20% (v/v) methanol for 15 min at room temperature. After staining, the plates were rinsed with H2O and air-dried.
Competitive growth assays
Cells were independently transduced with either a non-targeting sgRNA (co-expressed with either mCherry, GFP or BFP) or a targeting sgRNA. After 24 h, the cells were treated with the appropriate antibiotic, either puromycin (10 μg ml−1) or G418 (1.25 mg ml−1), until untransduced cells were selected from the population. The cells were then mixed at a 1:1 ratio by number and seeded in triplicate on six-well plates. After 24 h, cell populations were analysed using flow cytometry. The cells were either treated or not with exogenous agents, as indicated in the figures, and monitored using flow cytometry at the indicated time points thereafter.
Drug treatments
The chemicals used in this study can be found in Supplementary Table 7. Treatment durations and doses are indicated in the figures/legends.
Reverse transcription–quantitative PCR
One million RPE-1 cells were collected, and total RNA was isolated using the RNeasy Kits (QIAGEN), according to the manufacturer’s instructions. RNA was reverse transcribed into cDNA using iScript Reverse Transcription Supermix (Bio-Rad) using oligo(dT) primers. Quantitative PCR (qPCR) reactions were prepared with the SsoAdvanced Universal SYBR Green Supermix (Bio-Rad) and ran on a QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems). Primers used for qPCR can be found in Supplementary Table 7. Data were analysed using the Delta Delta Ct (ΔΔCt) method.
Nucleofection of RNase H1 constructs
For transient expression of RNase H1 constructs, 100,000 RPE-1 cells were transfected with 1 μg of RNase H1 plasmid using the Lonza Amaxa nucleofector with P3 solution and program EA104. Nucleofected cells were recovered in a warm medium and identified by mCherry expression using flow cytometry. All experiments with RNase H1 variants were carried out by gating only for the mCherry-positive cells.
Cell cycle analysis
The hTERT RPE-1 TP53 KO cells were arrested in G0/G1 by contact inhibition for 48 h and then re-plated at low density (100,000 cells per well on a six-well plate) to release them into S phase in the presence or absence of ML323 (30 μM). The cells were collected at the indicated time points, fixed in 70% ethanol for 30 min on ice, washed once with staining buffer (1× PBS + 5% FBS) and stained at room temperature using 1 µg ml−1 of 4′,6-diamidino-2-phenylindole (DAPI) solution (BD Biosciences) diluted in staining buffer for 5 min. For EdU–DAPI assay, 100,000 cells per well were seeded in a medium containing 1 µg ml−1 DOX. On the following day, the cells were treated with KSQ-4279 (25 μM) for 48 h and incubated with 10 μM EdU for the last 1 h before collecting. Click-iT Plus EdU Alexa Fluor 488 Flow Cytometry Assay Kit (Thermo Fisher Scientific) was used according to the manufacturer’s instructions, and the cells were resuspended in 1 µg ml−1 DAPI solution (BD Biosciences) diluted in 1× Click-iT permeabilization and wash reagent for 5 min. The cells were analysed using an Attune NxT Flow Cytometer. Subsequent analysis was performed using FlowJo v.10.8.1.
Western blotting
Cells were lysed in radioimmunoprecipitation assay buffer (0.5 M Tris-HCl (pH 7.4), 1.5 M NaCl, 2.5% deoxycholic acid, 10% NP-40 and 10 mM EDTA), supplemented with Halt Protease Inhibitor Cocktail and Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific). Samples were sonicated using a Bioruptor Plus sonicator (30 s ON and 30 s OFF for five cycles) and centrifuged for 5 min at 21,000g. Protein concentrations were measured using Bradford assay. The samples were normalized by protein concentration, mixed with 4× NuPAGE LDS Sample Buffer supplemented with 5% β-mercaptoethanol and boiled for 5 min at 98 °C. The samples were loaded into Bolt 4–12% Bis-Tris Protein Gels (Thermo Fisher Scientific) and transferred to 0.2-μm nitrocellulose membranes. Membrane blocking was performed with 5% milk diluted in Tris-buffered saline containing 0.1% Tween 20. Primary antibodies were diluted in 5% bovine serum albumin (BSA) diluted in Tris-buffered saline containing 0.1% Tween 20. After incubation with primary antibodies, membranes were incubated with Li-Cor near-infrared fluorescence or HRP secondary antibodies, after which they were scanned using a Li-Cor Near-InfraRed fluorescence Odyssey CLx Imaging System or FujiFilm LAS 4000 Gel Imager. All antibodies used are listed in Supplementary Table 7. All primary antibodies were used at a dilution of 1/1,000. All secondary antibodies were used at a dilution of 1/10,000.
Immunohistochemistry
Cells were seeded 48 h before on sterile 15-mm glass coverslips. The coverslips were washed in ice-cold PBS, and nuclei were 5 min pre-extracted by incubating cells in a pre-extraction buffer (HEPES (pH 7.5; 25 mM), NaCl (50 mM), EDTA (1 mM), MgCl2 (3 mM), sucrose (300 mM) and Triton X-100 (0.5%)) on ice. The cells were washed once in PBS and then fixed in 4% formaldehyde for 15 min at room temperature. The coverslips were again washed two times in PBS before blocking in 5% BSA in PBS ON at 4 °C. Primary antibody incubation with 53BP1 antibody (NB100-304) was carried out ON at 4 °C. After three washes in PBS, the coverslips were incubated with the secondary Alexa Fluor 488 antibody for 1 h at room temperature. The coverslips were washed three times in PBS and then incubated with Hoechst 33342 Ready Flow Reagent for 5 min before two more wash steps in PBS and a Milli-Q water wash. Cells were mounted onto glass slides using ProLong Diamond Antifade Mountant. All antibodies were diluted in 2.5% BSA in PBS. All incubation steps were carried out in a wet chamber. Images were taken using a Leica SP8 confocal microscope or ZEISS Apotome 3. For foci analysis, the layer with the highest intensity was selected. If there were foci in several planes, z-stack and maximum intensity projection were performed. For quantification of mitotic catastrophes, Hoechst staining was used to detect nuclear morphology, and fragmented nuclei were scored, as shown in Fig. 3e. The data were quantified using Fiji ImageJ v.2.9.0.
To detect 53BP1 nuclear bodies, cells were grown on a chambered coverslip for 24 h before being washed in PBS and fixed in 4% formaldehyde for 10 min at room temperature. The cells were washed three times in PBS and permeabilized with 0.5% Triton X-100 in cytoskeleton buffer (100 mM NaCl, 20 mM HEPES (pH 7.0), 3 mM MgCl2 and 300 mM sucrose) for 10 min at 4 °C. The cells were washed three times with PBS and blocked with 5% BSA (diluted in PBS) for 30 min. The cells were then incubated with primary antibodies (cyclin A, 1:200; 53BP1, 1:2,000) for 90 min at room temperature, washed three times in PBS and then stained with secondary antibodies (both 1:1,000) for an extra 45 min. ProLong Gold mounting reagent with DAPI was added to each coverslip. The cells were imaged using a Leica SP8 confocal microscope or ZEISS Apotome 3. Nuclear bodies in cyclin A-negative cells were manually counted. Representative images were prepared using QuickFigures ImageJ plugin70.
To detect ssDNA using BrdU labelling under native conditions, cells were grown on a chambered coverslip, incubated with 10 μM BrdU for 72 h and then pre-extracted, as described above. The cells were incubated with primary antibodies (BrdU, 1:250; RPA32, 1:500) ON and then stained with secondary antibodies (both 1:1,000) for 45 min. An ibidi mounting medium containing DAPI was used, and cells were imaged using a ZEISS Apotome 3. BrdU- and RPA32-positive foci were considered ssDNA stretches and were manually counted.
Metaphase spreads
Cells were treated with 0.04 μg ml−1 of colcemid for 16 h to arrest them in mitosis and then collected, washed with PBS and incubated in 0.075 M KCl at 37 °C for 10 min. Cell pellets were fixed in methanol:acetic acid (3:1 ratio), spread on glass slides and then coated with ProLong Diamond Antifade Mountant. Spreads were imaged using a ZEISS Apotome 3 microscope.
S1 nuclease DNA fibre assays
The S1 nuclease DNA fibre assay was conducted, as described in a previous study71, with the following modifications. Cells were incubated with 19 mM CldU for 20 min, washed three times with warm PBS and then incubated with 28 mM IdU for 60 min. During the IdU incubation, the cells were either treated or not with KSQ-4279 (25 μM). The cells were then collected and incubated with cytoskeleton-100 buffer (100 mM NaCl, 20 mM HEPES, 3 mM MgCl2, 300 mM sucrose and 0.5% Triton X-100) for 5 min and then washed with PBS. The cells were then incubated in 200-μl S1 nuclease reaction buffer with or without S1 nuclease (20 U ml−1; Thermo Fisher Scientific) for 30 min at 37 °C. The cells were then washed with PBS, lysed on glass slides in a buffer composed of 200 mM Tris-HCl (pH 7.5), 50 mM EDTA and 0.5% sodium dodecyl sulfate (SDS) for 5 min, and spread by tilting the slides at a 45° angle. The slides were then air dried and fixed in methanol:acetic acid (3:1 ratio) overnight at 4 °C. The slides were then incubated in 2.5 M HCl for 1 h to denature the DNA fibres, washed with PBS and incubated in 1% BSA/PBS (0.2% Tween 20) for 40 min. Staining was performed by incubating the slides for 2.5 h at room temperature with anti-CldU (1:500; ab6326; Abcam) and IdU (1:100; B44; 347,580; BD Biosciences) antibodies, followed by 1-h room temperature incubation with anti-mouse Alexa Fluor 488 (1:300) and anti-rat Alexa Fluor 568 (1:150) secondary antibodies. All antibodies were diluted in 2.5% BSA/PBS. Fibres were visualized using a Leica SP8 confocal microscope or ZEISS Apotome 3 microscope (×64; oil) and analysed using Fiji ImageJ v.2.9.0.
Live imaging
Cells were grown on chambered coverslips. PIP–FUCCI reporter-expressing cells were seeded into a medium containing 50–100 nM palbociclib. Twenty-four hours later, the medium was replaced with Dulbecco’s modified Eagle’s medium (no phenol red) supplemented with either dimethyl sulfoxide (DMSO) or KSQ-4279 (20 μM). For H2B–GFP experiments, cells were treated with DMSO or KSQ-4279 (25 μM) for 24 h before being imaged for an extra 24 h. All live cell imaging experiments were performed using a Nikon spinning disk equipped with a Yokogawa Confocal Scanner Unit CSU-W1-T2 SoRa at ×20 magnification.
ChIP–seq
For each condition, 10–20 million cells were fixed in 1% formaldehyde at room temperature for 15 min. The fixation reaction was quenched with glycine to a final concentration of 125 mM. Cells were collected and washed twice with chilled PBS, and pellets were snap frozen in a dry ice–ethanol bath before storing at −80 °C. When ready to process, cell pellets were thawed on ice and incubated with lysis buffer (LB)1 (50 mM HEPES–KOH (pH 7.5), 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40 or Igepal CA-630, 0.25% Triton X-100 and 1× protease inhibitors) on ice for 10 min. Cells were pelleted by centrifugation and incubated with LB2 (10 mM Tris-HCl (pH 8.0), 200 mM NaCl, 1 and 0.5 mM EDTA and 1× protease inhibitors) for 5 min on ice. The extracted nuclei were pelleted by centrifugation and resuspended in LB3 (10 mM Tris-HCl (pH 8.0), 100 mM NaCl, 1 mM EDTA, 0.1% Na deoxycholate, 0.5% N-lauroylsarcosine and 1× protease inhibitors). Nuclei were sonicated using a Covaris S2 sonicator with the following settings: duty cycle 5%, intensity 5, 200 cycles per burst and 12 min. Debris was pelleted by centrifugation at 4 °C, and the supernatant was transferred to a 5-ml tube. Then, 100 µl of Dynabeads Protein A that had been prebound with MRE11 (catalogue no. NB100-142; Novus), FANCM, SMARCAL1 or DNA cruciform antibody was added to the cell lysate, and samples were incubated at 4 °C overnight with rotation. Beads were collected on a magnetic stand and washed with ice-cold radioimmunoprecipitation assay buffer six times, followed by a final wash with Tris-buffered saline before resuspending beads in 200 µl of elution buffer (50 mM Tris-HCl (pH 8.0), 10 mM EDTA and 1% SDS). The bead slurries were incubated overnight at 65 °C to reverse crosslinks. Samples were treated with 1 mg ml−1 RNaseA (catalogue no. 2271; Ambion) for 30 min at 37 °C, followed by proteinase K treatment (20 mg ml−1; catalogue no. 25530-049; Invitrogen) for 1 h at 55 °C. DNA was then purified using a MinElute PCR Purification Kit (catalogue no. 28004; QIAGEN), and sequencing libraries were prepared using a NEBNext Ultra II kit.
ChIP–seq analysis
Raw paired-end FASTQ files were aligned to the GRCh38 reference genome using Bowtie 2 v.2.4.4. The resulting BAM files were sorted and indexed with Samtools v.1.6. Peaks were called using MACS3 v.3.0.0b1 with RPE-1 wild type set as the control and default settings. Peaks were filtered using the ENCODE blacklist v2 and manually inspected72. Total read counts were calculated for each sample and used to generate comparable bigWig files for visualization using bamCoverage v.3.3.0 and ScaleFactor option. CrossMap v.0.6.0 was used to lift over GRCh38 to GRCh19 coordinates. Locally hosted bigWig files were added as custom tracks to the UCSC Genome Browser for visual inspection. MEME-ChIP v.5.5.1 was used to identify motifs around the peaks, and ChIPseeker was used to visualize peak locations over chromosomes. The AT fraction was calculated per base across all peaks in a 1,000-bp window around the centre of each peak. In Fig. 4e, MRE11 enrichment was determined at a union set of TA repeat sites (more than 70% TA content) consisting of FANCM peaks in SMARCAL1 KO cells, SMARCAL1 peaks in FANCM KO cells and unique cruciform and MRE11 peaks in double-perturbed cells.
Repli-seq analysis
Repli-seq data for GM06990 cells from the University of Washington Encyclopedia of DNA Elements group were downloaded from the UCSC Genome Browser as bigWig files in wavelet-smoothed and percentage-normalized signal format (GEO accession: GSM923443). DeepTools v.3.5.1 was used to visualize the wavelet-smoothed signal data for all peaks in a 2-Mb region around each peak centre with the tools computeMatrix reference-point, the bin-size option set to 1 and plotHeatmap to create the heat map. R was used to process and plot the percentage-normalized signal data.
Protein purification
Human SMARCAL1
The hSMARCAL1 wild-type and ΔRBM variants were expressed in Spodoptera frugiperda 9 (Sf9) cells using the Bac-to-Bac expression system (Thermo Fisher Scientific), according to the manufacturer’s recommendations. The protein sequences were codon optimized for the expression in Sf9 insect cells and cloned NheI and XhoI restriction sites of pFastBac1, generating pFB-2xMBP-hSMARCAL1-10xHis. Frozen Sf9 pellets from 200-ml cultures for each variant were resuspended in LB (50 mM Tris-HCl (pH 7.5), 5 mM β-mercaptoethanol, 1 mM phenylmethanesulfonylfluoride (PMSF), 1 mM EDTA, protease inhibitor cocktail (P8340; Sigma-Aldrich) diluted 1:300 and 30 µg ml−1 of leupeptin (Merck Millipore)) and incubated at 4 °C for 20 min. Glycerol was added to a final concentration of 16.7%. NaCl was added to a final concentration of 305 mM. The solution was incubated at 4 °C for 30 min. The mixture was centrifuged at 20,000g at 4 °C for 30 min. The resulting soluble extract was incubated with 4-ml amylose resin (NEB) at 4 °C for 1 h. The resin was washed with amylose wash buffer 1 M (50 mM Tris-HCl (pH 7.5), 5 mM β-mercaptoethanol, 1 mM PMSF, 10% glycerol and 1 M NaCl) followed by 300 mM amylose wash buffer (50 mM Tris-HCl (pH 7.5), 5 mM β-mercaptoethanol, 1 mM PMSF, 10% glycerol and 300 mM NaCl). Proteins were eluted using amylose elution buffer (50 mM Tris-HCl (pH 7.5), 5 mM β-mercaptoethanol, 1 mM PMSF, 10% glycerol, 300 mM NaCl and 10 mM maltose). The MBP-tagged variants were incubated with PreScission Protease (approximately 10 μg of PreScission protease per 100 μg of tagged protein) at 4 °C for 1 h to cleave the MBP tag. Subsequently, the cleaved protein was diluted 1:3 to reduce the concentration of NaCl to approximately 100 mM with a buffer containing no NaCl (50 mM Tris-HCl (pH 7.5), 5 mM β-mercaptoethanol, 1 mM PMSF and 10% glycerol) and applied in flow on a column containing pre-equilibrated CM Sepharose resin (GE HealthCare). The resin was washed with Sepharose wash buffer 100 mM NaCl (50 mM Tris-HCl (pH 7.5), 5 mM β-mercaptoethanol, 1 mM PMSF, 10% glycerol and 100 mM NaCl). SMARCAL1 variants were eluted with Sepharose buffer 300 mM NaCl (50 mM Tris-HCl (pH 7.5), 5 mM β-mercaptoethanol, 1 mM PMSF, 10% glycerol and 300 mM NaCl). Fractions containing high protein concentration were pooled, aliquoted, snap-frozen in liquid nitrogen and stored at −80 °C.
Human FANCM
FANCM was expressed in S. frugiperda 9 (Sf9) cells using the Bac-to-Bac expression system (Thermo Fisher Scientific), according to the manufacturer’s recommendations. The protein sequence was codon optimized for the expression in Sf9 insect cells (Twist Bioscience) and cloned into NheI and AflII restriction sites of pFastBac1, generating pFB-2xMBP-hFANCM-10xHis. Frozen Sf9 pellet from 1.5-l culture was resuspended in LB (50 mM Tris-HCl (pH 7.5), 5 mM β-mercaptoethanol, 2 mM PMSF, 1 mM EDTA, protease inhibitor cocktail (P8340; Sigma-Aldrich) diluted 1:200 and 50 µg ml−1 of leupeptin (Merck Millipore)) and incubated at 4 °C for 5 min. Glycerol was added to a final concentration of 16.7%. NaCl was added to a final concentration of 305 mM. The solution was incubated at 4 °C for 30 min. The mixture was centrifuged at 20,000g at 4 °C for 30 min. The resulting soluble extract was incubated with 24-ml amylose resin (NEB) at 4 °C for 1 h. The resin was then washed with 1 M amylose wash buffer (50 mM Tris-HCl (pH 7.5), 5 mM β-mercaptoethanol, 1 mM PMSF, 10% glycerol, 1 mM EDTA and 1 M NaCl) followed by 300 mM amylose wash buffer (50 mM Tris-HCl (pH 7.5), 5 mM β-mercaptoethanol, 1 mM PMSF, 10% glycerol and 300 mM NaCl) and 150 mM amylose wash buffer (50 mM Tris-HCl (pH 7.5), 5 mM β-mercaptoethanol, 1 mM PMSF, 10% glycerol and 150 mM NaCl). Proteins were eluted using amylose elution buffer (50 mM Tris-HCl (pH 7.5), 5 mM β-mercaptoethanol, 1 mM PMSF, 10% glycerol, 150 mM NaCl and 10 mM maltose). The MBP-tagged variants were incubated with PreScission Protease (approximately 20 μg of PreScission Protease per 100 μg of tagged protein) at 4 °C for 1 h to cleave the MBP tag. Subsequently, the cleaved protein was diluted to reduce the concentration of NaCl to approximately 100 mM with a buffer containing no NaCl (50 mM Tris-HCl (pH 7.5), 5 mM β-mercaptoethanol, 1 mM PMSF and 10% glycerol) and applied in flow on a column containing pre-equilibrated HiTrap Q HP column on the ÄKTA purifier (Cytiva). The protein was washed with 20 ml of ÄKTA buffer (50 mM Tris-HCl (pH 7.5), 5 mM β-mercaptoethanol, 1 mM PMSF and 10% glycerol) at 100 mM NaCl and eluted from the column with a salt gradient in ÄKTA buffer from 100 mM to 1 M NaCl. Fractions containing the protein were pooled, aliquoted and snap-frozen.
Secondary structure unfolding assays
Secondary structure unfolding assays (15 μl) were carried out in a reaction buffer containing 25 mM of Tris-acetate (pH 7.5), 2 mM ATP, 2 mM magnesium acetate, 1 mM dithiothreitol, 0.1 mg ml−1 Recombinant albumin (NEB) and 100 ng of DNA substrate per reaction (pUC19, pUC19_TA rich or pUC19_Chr 3). The final NaCl concentration was adjusted to 50 mM accounting for the salt brought into the reactions with protein storage or protein dilution buffer. The reactions were assembled and supplemented with the relevant proteins (with and without 20 nM RPA, as indicated) on ice and then incubated at 37 °C for 30 min. The reactions were then incubated with T7 Endonuclease I (NEB) or EcoRI (NEB) for 10 min at 37 °C, and where indicated, followed by SspI (NEB) for 30 min at 37 °C. The reactions were terminated by adding 1-μl Proteinase K (14–22 mg ml−1; Roche) and 5-μl 0.2% stop solution (150 mM EDTA, 0.2% SDS, 30% glycerol and bromophenol blue) and incubated at 37 °C for 10 min. The products were separated by electrophoresis in 1% native agarose gels in the presence of GelRed (1:20,000; Biotium). Images of gels were acquired using Quantum (CX5 Edge; VILBER). The images were quantified using ImageJ and expressed as percentage of secondary structure removal.
DNA substrate preparation
The pUC19_Chr3 vector was obtained by inserting a representative sequence (5′-TGTATATATATACAATATATATACATGTATATATATACATGTATATATACTGTATATATACATGTATATATATACATGTATATATACTGTATATATACAT-3′) into the SacI and XbaI restriction sites of a pUC19 vector. The selected sequence from chromosome 3 was found in ChIP–seq experiments and was predicted to be a substrate for FANCM, SMARCAL1 and MRE11. The pUC19_TA rich vector instead contains (TA)20 inserted into the KpnI and BamHI restriction sites of pUC19 (ref. 73).
Liquid chromatography–tandem mass spectrometry sample preparation
Proteins were extracted in 150-μl LB (4% SDS and 50 mM Tris-HCl (pH 8.2)) using a tissue homogenizer (TissueLyser II; QIAGEN) with glass beads and 2 × 2 min cycles at 30 Hz. The samples were treated with high-intensity focused ultrasound (HIFU) for 1 min at an ultrasonic amplitude of 100% before boiling at 95 °C for 5 min while shaking at 800 rpm on a thermoshaker (Eppendorf). Cell debris and other insoluble components were separated by centrifugation at 20,000g for 10 min. Protein concentration was determined using the Lunatic UV/Vis polychromatic spectrophotometer (Unchained Labs).
For each sample, a volume corresponding to 780 µg of protein was taken and supplemented with 5 mM tris(2-carboxyethyl)phosphine and 15 mM chloroacetamide at 30 °C for 30 min in the dark.
Samples were processed using the single‐pot solid‐phase-enhanced sample preparation. Protein purification, digest and peptide clean-up were performed using a KingFisher Flex System (Thermo Fisher Scientific) and Carboxylate-Modified Magnetic Particles (GE65152105050250 and GE45152105050250; GE Life Sciences)74. Samples were diluted with 100% ethanol to a final concentration of 60% ethanol. The beads, wash solutions (80% ethanol) and samples were loaded into 96-deep-well or microplates and transferred to the KingFisher. Collection of beads, protein binding to beads, washing of beads, protein digestion (overnight at 37 °C with a trypsin:protein ratio of 1:50 in 50 mM triethylammonium bicarbonate) and peptide elution were carried out on the robotic system. The digest solution and water elution were combined and dried to completeness.
Antibody-based enrichment of the ubiquitin remnant motif (K-ε-GG) was performed according to the manufacturer’s protocol (PTMScan; Cell Signaling Technology), except for using only 40% of the recommended bead volume. Both eluates were loaded directly onto a preconditioned Evotip (Evosep Biosystems) according to the manufacturer’s protocol.
Liquid chromatography–tandem mass spectrometry data acquisition (proteomics)
Mass spectrometry analysis of proteomics samples was performed on a timsTOF Pro (Bruker) coupled to an Evosep One (Evosep Biosystems). Samples were separated using the 15 samples per day method while keeping the analytical column (PepSep C18, 15 cm × 150 µm; 1.5 µm) at 50 °C. Mass spectrometry scans were acquired from m/z 100 to m/z 1,700 with an inverse mobility ramp (1/K0), where K0 represents the reduced ion mobility, from 0.60 to 1.60 Vs cm−2 (ion accumulation and ramp time both set at 100 ms). MS2 scans were acquired in data-independent acquisition parallel accumulation–serial fragmentation mode. One mass spectrometry scan was followed by 16 parallel accumulation–serial fragmentation cycles from m/z 400 to m/z 1,200, with overlapping isolation windows of m/z 26, covering 2 × 0.30 (1/K0) ion mobility windows in the range of 0.60–1.42 Vs cm−2. Singly charged ions were excluded using the ion mobility polygon filter mask. The mass spectrometry proteomics data were handled using the local laboratory information management system B-Fabric75.
Protein identification and quantification
The acquired mass spectrometry data were processed for identification and quantification using Spectronaut (v.19.0240606.62635; Biognosys) in directDIA mode. Spectra were searched against a canonical Swiss-Prot database for human and common protein contaminants (NCBI taxonomy ID 9606; release date 30 March 2023). Carbamidomethylation of cysteine was set as fixed modification, whereas methionine oxidation, N-terminal protein acetylation, GlyGly and LeuArgGlyGly on lysine residues were set as variable modifications. Enzyme specificity was set to trypsin/P allowing a minimal peptide length of seven amino acids and a maximum of two missed cleavages. Precursor and fragment tolerance was set to dynamic for the initial search. The maximum false discovery rate was set to 0.01 for peptides and 0.01 for proteins. Protein quantification was performed in Spectronaut using the default post-translational modification settings. The quantitative data were extracted using the BGS Factory Report (default) and used for follow-up analyses.
For statistical evaluation and site-specific integration of the results, the Spectronaut modification output was filtered for only confidently assigned diGly modified peptides, and for each diGly peptide assignment, the number of diGly modifications, the modified residue and position in the protein were parsed into a data frame. Further, a new identifier was generated that comprised the protein accession, the diGly acceptor residue and the diGly site in the full-length protein. This identifier was then used to summarize (sum) quantities from each sample. This site-centric data are then further analysed using the prolfqua R package76.
For this, the data are log2-transformed and normalized with a robust z-score transformation. A linear model is fitted to each diGly peptide abundance to compute the significant differences between different conditions. These group comparisons (contrasts) were evaluated with a moderated Wald test with pooled variance (as implemented in the limma R package77. The resulting P values were adjusted for multiple testing using the Benjamini–Hochberg method. All relevant mass spectrometry data are included in the Source data Excel file.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.