Bacterial and fungal culture conditions
Fungal strains, bacterial strains and plasmids used in this study are listed in Supplementary Tables 14 and 15. S. Typhimurium strains were grown at 37 °C in lysogeny broth (LB) medium (per litre: 10 g tryptone, 5 g yeast extract and 10 g sodium chloride). C. albicans strains were grown at 37 °C in YPD media (per litre: 10 g yeast extract, 20 g peptone and 20 g dextrose). Antibiotics and antifungals were added at the following concentrations to LB agar or YPD agar, as needed: 100 mg l−1 carbenicillin–ampicillin, 30 mg l−1 chloramphenicol, 50 mg l−1 kanamycin, 1 or 10 mg l−1 doxycycline, 2.5 mg l−1 amphotericin, 200 or 25 mg l−1 nourseothricin (NAT) and 600 pr 75 mg l−1 hygromycin B. All strains were cultured aerobically shaking at 200 rpm overnight except for in vitro experiments where Salmonella strains were grown overnight in liquid LB without shaking at 37 °C. Co-incubation of S. Typhimurium and C. albicans strains were done by quantifying the cell number by measuring the optical density at 600 nm (OD600) of the cultures. Cells of Salmonella strains (1 × 109) and cells of C. albicans strains (1 × 108) were centrifuged and resuspended in 1 ml LB and co-incubated for 2 h in 37 °C without shaking, unless indicated otherwise. The cell-free supernatant from the 2 h co-incubations was filter sterilized and used to co-incubate with Salmonella to analyse the effect of secreted factors. When indicated, 1 × 108 cells of C. albicans were heat killed at 100 °C for 1 h, before co-incubation with Salmonella.
Animal models
Specific pathogen-free mice
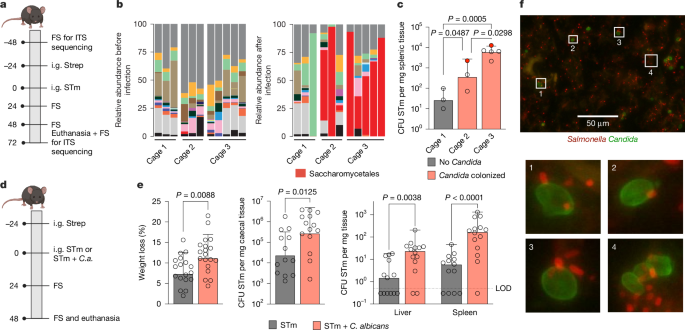
All animal experiments were reviewed and approved by the Institutional Animal Care and Use Committee at the University of Illinois Chicago in protocols 17-045, 20-016 and 22-192 and were in agreement with ethical regulations. Eight-to-nine-week-old C57BL/6J female or male WT mice (strain number 000664) and CBA/J female WT mice (strain number 000656) were obtained from Jackson Laboratories, maximum barrier, and were 9–10 weeks of age by the start of every experiment. Mice were housed in the Biologic Resources Laboratory facility at the University of Illinois Chicago in individual cages with filter tops (maximum occupancy of five mice) containing corn cob bedding and paper nesting material. The room the mice were housed in was kept under 14 h–10 h light–dark cycles, 70–76 °F and 30–70% humidity. Mice had access to food and autoclaved water ad libitum. Mice were fed chow LM485. Mice were randomized to experimental cages before the experiments. Experimenters were not blinded to experimental details in mouse experiments. No sample-size calculation was performed. Sample sizes were chosen based on past experience using the methods previously described61,62,63,64. Mouse experiments were performed with 2–6 mice per group and repeated for a minimal number of 5 mice per experimental condition. For infections with C57BL/6J mice, mice were pre-treated with streptomycin (0.1 ml of a 200 mg ml−1 solution in sterile water) intragastrically 24 h before inoculation with Salmonella (WT) or C. albicans (WT) or Salmonella (ΔsopB or ΔSTM4351) and C. albicans (WT, arg4Δ/Δ or arg4Δ/Δ + ARG4). For initial experiments, 1 × 109 colony-forming units (CFU) ml−1 of Salmonella and 1 × 108 CFU ml−1 of C. albicans were used (Figs. 1a–e and 5a,b and Extended Data Figs. 1c–e, 7a,c–e and 8a,c,d). The same dose, however, yielded significantly higher colonization and dissemination of Salmonella in single infected mice in subsequent experiments. We therefore maintained the same ratio of Salmonella to C. albicans but decreased the overall dose to 1 × 107 CFU ml−1 of Salmonella and 1 × 106 CFU ml−1 of C. albicans. This dose yielded Salmonella caecal colonization and dissemination levels similar to previous experiments (Figs. 2i, 3c,d, 4f and 5c–k and Extended Data Figs. 3h,i, 4f–i, 7b, 8b,e–h and 9d–i). For experiments with C. albicans arginine auxotroph (arg4Δ/Δ), mice were either gavaged again with C. albicans at 24 h post-infection or the day 0 dose of C. albicans was increased to 1 × 107 CFU ml−1 (Figs. 3c,d and 4f and Extended Data Figs. 1f, 4f–h and 6f–i) and excluded the mice if C. albicans did not colonize in the caecum. When required, mice were treated with 2% l-arginine (adjusted to approximately pH 7) or 20 mM l-lysine (adjusted to approximately pH 7) in their drinking water ad libitum. For infections with CBA/J mice, mice were colonized intragastrically with 1 × 108 CFU ml−1 of C. albicans 529L or PBS control 3 and 1 days before and, to maintain equal colonization levels across mice, 4 days post intragastrically infection with 1 × 109 CFU ml−1 of Salmonella. Inflammation develops slower in this model, with peak inflammation reached at 9–10 days post-infection65. Weights were monitored daily for both mouse models. Faecal samples were collected daily, and serial tenfold dilutions were plated for enumerating bacterial CFU on LB agar plates supplemented with either carbenicillin or kanamycin with amphotericin or YPD agar plates with chloramphenicol. Mice were euthanized at time points indicated. Caecal colonization and dissemination were assessed by homogenization and plating of the caecum, spleen, liver, Peyer’s patches and mesenteric lymph nodes, respectively. Colon content was collected to enumerate Salmonella colonization and was flash frozen for microbiota analysis. Caecal luminal content was also collected, and flash frozen to either isolate RNA or perform metabolomic analysis. Caecal tissue was collected by flash freezing to analyse mouse inflammatory gene expression and by fixing it with formalin for histological examination. No statistical methods were used to pre-determine sample sizes, but our sample sizes are similar to those reported in previous publications62,63,64.
Gnotobiotic mice
Swiss Webster (Tac:SW) germ-free WT mice were purchased from Taconic and C57BL/6 mice were purchased from Jax and bred at the BRL facility at the University of Illinois Chicago in a room with 14 h–10 h light–dark cycles and 70–76 °F and 30–70% humidity. C57BL/6 germ-free mice for some experiments were obtained from the University of Illinois Chicago Gnotobiotic core. Mice were kept in isolators purchased from Park Bioservices. Mice were fed autoclaved 5L79 chow and autoclaved super Q water ad libitum. Germ-free conditions were tested at least once a month with aerobic liquid cultures (brain heart infusion), solid cultures (blood agar plates; R01200, Thermo Sci Remel), fungal cultures (Sabouraud slants), anaerobic liquid (brain heart infusion) and solid cultures (Brucella agar; R01254, Thermo Sci Remel) from swabs of isolators, faecal samples and fungal traps placed inside the isolators. Faecal samples were also tested with Gram staining and qPCR to detect bacterial DNA. For ASF experiments, mice were stably colonized with ASF purchased from Taconic Biosciences. Stable colonization was assessed via species-specific PCR66.
For immunofluorescence and Candida gene expression experiments, 10–15-week-old ASF-colonized Swiss Webster male and female mice were housed in a biosafety cabinet. Mice were then intragastrically infected with 0.1 ml of 1 × 107 CFU ml−1 of Salmonella or 1 × 107 CFU ml−1 of Salmonella and 1 × 106 CFU ml−1 of C. albicans. Faecal samples were collected, and serial tenfold dilutions were plated for enumerating bacterial CFU on LB agar plates supplemented with carbenicillin and amphotericin or YPD agar plates with chloramphenicol. Mice were euthanized at 6 h and 24 h post-infection and caecal and colonic samples were collected by fixing it with formalin for immunofluorescence. For Candida gene expression experiments, mice were euthanized at 6 h and 12 h and luminal caecal content samples were collected and snap frozen.
For germ-free mouse experiments, 8–10-week-old C57BL/6 mice were housed in a biosafety cabinet. Mice were inoculated with 1 × 107 CFU ml−1 of C. albicans intragastrically 24 h before infecting with 1 × 104 CFU ml−1 of Salmonella. Mice were euthanized at 24 h post-Salmonella infection. For ASF mouse experiments, 8–10-week-old ASF-colonized C57BL/6 mice were housed in a biosafety cabinet. Mice were inoculated with 1 × 107 CFU ml−1 of C. albicans intragastrically 24 h before infecting with 1 × 107 CFU ml−1 of Salmonella. Mice were euthanized at 48 h post-Salmonella infection. Faecal samples were collected, and serial tenfold dilutions were plated for enumerating bacterial CFU on LB agar plates supplemented with carbenicillin and amphotericin or YPD agar plates with chloramphenicol. Caecal colonization and dissemination were assessed by homogenization and plating of the caecum, spleen and liver, respectively. Colonic content was collected to enumerate Salmonella colonization. Caecal luminal content was also collected, and flash frozen to either isolate RNA or perform metabolomic analysis. Caecal tissue was collected by flash freezing to analyse mouse inflammatory gene expression and by fixing it with formalin for histological examination.
Both male and female mice were used in this study. Most experiments were conducted using female mice, with the repeat of key experiments in male mice to ensure reproducibility across sexes. Specifically, Fig. 1f includes five male and three female mice per group. Extended Data Fig. 5a–d includes five female and two male mice in the STm group and six female and two male mice in the STm + C. albicans group. Extended Data Fig. 5e–h includes three male and two female mice per group.
Mycobiota and microbiota sequencing and analysis
ITS1 and 16S sequencing of mouse faecal samples were performed largely as previously described12,61. In brief, for DNA isolation, one to two mouse faecal pellets were resuspended in 0.5 ml lyticase buffer (50 mM Tris, 1 mM EDTA and 0.2% 2-mercaptoethanol), homogenized briefly and treated with 200 U lyticase (Sigma-Aldrich) for 30 min at 30 °C. Material was pelleted and resuspended in 0.4 ml stool DNA stabilizer (B Bridge International), mixed with 0.1 ml 0.1-mm silica beads (Biospec) and 0.3 ml 0.5-mm beads (Biospec), heated at 95 °C for 5 min and subjected to bead beating twice on high (VWR) for 1 min. DNA was then further purified using the QIAmp DNA mini kit (Qiagen) according to the manufacturer’s instructions.
Primers used in this study are detailed in Supplementary Table 16. Fungal ITS1 amplicons were generated using primers with the following recombinant DNA targeting sequences: ITS1 forward (5′-CTTGGTCATTTAGAGGAAGTAA) and ITS2 reverse (5′-GCTGCGTTCTTCATCGATGC). ITS1 amplicons were generated with 35 cycles using Invitrogen AccuPrime PCR reagents at an annealing temperature of 48 °C. Bacterial 16S (V1–V3 region) amplicons were generated using primers with the following recombinant DNA-targeting sequences: 27 forward (5′-AGAGTTTGATCMTGGCTCAG) and 534 reverse (5′- ATTACCGCGGCTGCTGG). Amplicons were then used in the second PCR, using Illumina Nextera XT v2 barcoded primers to uniquely index each sample, and 2 × 300 paired-end sequencing was performed on the Illumina MiSeq.
ITS1 sequences were trimmed using ITSxpress (v1.7.4)67 in QIIME 2 (v2019.7). Using the DADA2 package (v1.10.1) in R (v3.5.2), reads underwent further quality filtering as error rates were calculated and removed from the dereplicated reads. Where forward and reverse reads could be merged, they were, and where they could not, largely owing to ITS1 sequences sometimes being longer than sequence coverage, they were concatenated. An initial sequence table was constructed before chimeras were identified using the removeBimeraDenovo function. Finally, taxonomy was assigned using DADA2’s native naive Bayesian classifier against the UNITE (v.8.2) database68. 16S sequences were trimmed using Cutadapt (v3.7) and taxonomy was assigned with the Green-Genes reference database (release of May 2013).
Statistical analyses were performed with R (v4.0.2)69. Figures were produced using the packages ggplot2 (ref. 70), dplyr71, ape72 and RColorBrewer73. Microbial communities were further analysed using the microbiome74 and phyloseq75 packages.
Fluorescence microscopy
For in vivo experiments, tissue samples were fixed in 10% formalin for 24 h, stored in 70% ethanol and finally embedded in paraffin and sectioned at 7 μm with a microtome. Deparaffinization was performed by immersing the sections in xylene followed by decreasing concentrations of ethanol (100%, 90% and 70%). For antigen retrieval, the deparaffinized slides were immersed in 10 mM Na-citrate buffer and heated in a microwave for 20 min. Blocking was performed by flooding the slides with PBS–10% FBS for 30 min at 4 °C. Salmonella was stained with Salmonella antisera O-4 (1:100 in PBS; 294401, Hardy Diagnostics) for 1 h at room temperature. Following washing with PBS–0.3% Tween20, slides were incubated for 30 min at room temperature with Alexa Fluor 594 goat anti-rabbit antibody (1:1,000 in PBS; R37117, Invitrogen). Slides were washed with PBS–0.3% Tween20 and C. albicans was stained using a FITC-labelled rabbit anti-fungal antibody (1:250 in PBS–10% FBS; B65411F, Meridian Life Science) for 1 h at 37 °C. Slides were washed with PBS–0.3% Tween20, mounted using ProLong Diamond Antifade Mounting with DAPI (Invitrogen) and images were taken using the BZ-X710 All-in-One Fluorescence Microscope at ×60.
For in vitro experiments, to visualize agglutination, OD600 of overnight Salmonella–mCherry and C. albicans–GFP was measured. STm (4 × 109) and C. albicans (7.6 × 107) were centrifuged and resuspended in 1 ml and 2 ml of PBS, respectively. On a microscope slide, 20 µl of Salmonella and 20 µl of C. albicans were added to the slide and mixed by swirling the slide for 1 min. Of the mixture, 5 µl was added to an agarose pad (0.75 g of agarose in 50 ml of H2O). Images were taken using the BZ-X710 All-in-One Fluorescence Microscope at ×4–40. For quantification of prgH-gfp+ cells, 5 µl of 1 × 108 STm SB300 prgh-gfp cells (expressing mCherry) alone or with 1 × 108 C. albicans were spotted onto a 1 × 1-cm-wide, 3-mm-thick agarose pad (1.5% agarose in M9 minimal medium) positioned on a microscope slide and allowed to dry for 10 min. Once cells were absorbed onto agarose pads, coverslips were added, sealed with nail polish and then slides were incubated for 2 h at 37 °C. Cells were visualized at ×60 on a BZ-X710 All-in-One Fluorescence Microscope. The acquired images were not always completely in focus. Therefore, a section of each image in focus was selected for counting. Each selected section contained at least ten C. albicans cells. In each section, the number of mCherry-positive (total) and GFP-positive (Prgh-expressing) cells were counted and presented as the percentage of GFP+:mCherry+.
Invasion (gentamicin protection) assay
The T84 colonic epithelial cell line (CCL-248, ATCC; RRID:CVCL_0555) or C2BBe1 colonic epithelial cell line (Caco2; CRL-2102, ATCC; RRID:CVCL_1096) were obtained directly from the manufacturer, confirmed visually and tested for mycoplasma contamination. Cells were seeded onto a 24-well tissue-culture-treated plate at a density of 5 × 105 cells per well with media lacking antibiotics and/or antimycotics and incubated overnight (for T84) and for 5 days (for Caco2) at 37 °C. Salmonella strains alone or co-incubated with either C. albicans strains or the cell-free supernatant were centrifuged, resuspended in DMEM/F12 and serially diluted. Epithelial cells were infected with 5 × 105 cells of Salmonella (MOI = 1) and 5 × 104 cells of C. albicans (MOI = 0.1). Infected cells were incubated at 37 °C for 1 h. The inoculum was serially diluted and plated on LB agar with amphotericin to confirm bacterial numbers. After infection, the medium was removed via vacuum, and wells were washed three times with 500 μl PBS. Of DMEM/F12 + 10% FBS + 0.1 mg ml−1 gentamicin, 500 μl was added to the wells and incubated at 37 °C for 1 h to kill extracellular bacteria. After incubation, the wells were washed with PBS and lysed by incubation with 1% Triton X-100 for 5–10 min. Cells were disrupted and harvested by scraping wells and pipetting, were serially diluted and plated on LB agar with amphotericin to quantify bacterial cells that invaded. The percentage of cells recovered relative to the inoculum was calculated.
Sedimentation assay
OD600 of overnight Salmonella and C. albicans cultures was measured. Salmonella (4 × 109) and C. albicans (7.6 × 107) were centrifuged and resuspended in 1 ml and 2 ml of PBS, respectively. In a 15-ml conical tube, 1 ml of Salmonella and 2 ml of C. albicans were added and vortexed for 15 s before taking 200 μl from the top of the mixture, which was used to measure OD600 (1:5 dilution) for the initial starting value. For a baseline control, 2 ml of C. albicans was mixed with 1 ml of PBS and the OD600 was measured. Conical tubes (15 ml) were then incubated at 37 °C for 20 min without shaking; after that time, OD600 was measured the same way as before. To calculate the percentage of sedimentation, the equation ((OD0 min – OD20 min)/OD0 min) × 100 was used. The value of Salmonella and C. albicans was divided over the C. albicans alone to measure sedimentation.
RNA extraction
For the in vivo experiment, caecal tissue collected from mice 24 h post-infection and 48 h post-infection were homogenized by mortar and pestle using liquid nitrogen. Because mice were treated with streptomycin 24 h before infection, we collected caecal tissue from uninfected mice after streptomycin treatment as a control. The homogenate was transferred to 1 ml of Tri-Reagent (Molecular Research Center) for RNA extraction. RNA was extracted with 0.1 ml of bromo-3-chloropro-pane, centrifuged and the upper phase was precipitated with 0.5 ml isopropanol. After centrifugation, pellets were washed twice with 1 ml of 75% ethanol in RNase-free water. The RNA pellet was then resuspended in RNase-free water. For caecal content, RNA was extracted from snap-frozen luminal caecal content samples collected during mouse experiments. RNA extraction was performed using the Qiagen RNeasy Power Microbiome kit. RNA was treated with DNase using the Turbo DNA-free kit (Invitrogen). For the in vitro experiments, RNA extraction was performed on the cell pellet from monocultures and co-cultures of Salmonella and C. albicans after 2 h of co-incubation, except to assess the expression of sopB in C. albicans in the C. albicans tetO–sopB strain. Here transformants containing CIptet-SopB or CIpSATtetTranstADH1 (empty vector) were grown overnight in liquid YPD media containing 10 μg ml−1 doxycycline. Cells (1 × 105 ml−1) were transferred into 5 ml YPD with or without 10 μg ml−1 doxycycline and incubated for 8 h at 30 °C with continuous shaking. In all cases, RNA extraction was performed using the hot-phenol method as previously described76,77 followed by DNase treatment using the Turbo DNA-free kit (Thermo Fisher).
RNA sequencing and analysis
For basic processing of RNA sequencing data, raw reads were aligned to the reference assembly (GCF_000022165.1) using BWA-MEM (v0.7.17)78. Expression levels of gene features, that is, coding DNA sequences regions from the reference assembly, were quantitated using FeatureCounts (v2.0.3) as raw read counts of the stranded libraries79.Differential analysis of quantitated gene features compared with treatment was performed using the software package edgeR on raw sequence counts80. Before analysis, the data were subsampled to a maximum depth of 1,750,000 counts per sample and filtered to remove any features that had less than 100 total counts summed across all samples. Data were normalized as counts per million and an additional normalization factor was computed using the trimmed mean of M values algorithm. Statistical tests were performed using the ‘exactTest’ function in edgeR. Adjusted P values (q values) were calculated using the Benjamini–Hochberg false discovery rate (FDR) correction81. Significant gene features were determined based on an FDR threshold of 5% (0.05). For enrichment analysis, the enrichment or over-representation of differentially expressed gene features in the various gene groups, that is, pathways, modules and BRITE categories, listed for the KEGG organism ‘seo’ (KEGG genome T01714) was determined using Fisher’s exact test in R. In brief, a list of differentially expressed gene features was obtained from the results of the differential analysis based on a q value, that is, FDR-corrected P < 0.05. The enrichment of significantly different gene features as compared with all genes listed in KEGG for the organism ‘seo’ were then tested for the KEGG organism pathways, modules and BRITE level 1, 2 and 3 categories. Adjusted P values (q values) were calculated using the Benjamini–Hochberg FDR correction81. Significant enrichment of gene groups was determined based on an FDR threshold of 5% (0.05).
rt-qPCR
Reverse transcription was performed with the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). For Salmonella RNA, reactions were also performed without the addition of reverse transcriptase to confirm that there was no amplification of DNA in rt-qPCRs. Of RNA, 500 ng was used for the reverse transcription reaction. The reverse transcription cycle consisted of 10 min at 25 °C followed by 120 min at 37 °C and 5 min at 85 °C. rt-qPCR was performed using the Fast SYBR Green Master Mix (Applied Biosystems) on the Viia7 Real-time PCR system at the Genome Research core at the University of Illinois at Chicago. The rt-qPCR cycle consisted of 20 s at 95 °C followed by 40 cycles of 3 s at 95 °C and 30 s at 60 °C. Reactions were performed in duplicate.
For transcript levels of sopB in doxycycline-repressible strains, samples were normalized to 1 µg RNA and treated with Turbo DNase (Thermo Fisher). First-strand cDNA synthesis was performed using the RevertAid RT kit following the manufacturer’s protocol. Amplification of approximately 20 ng cDNA was performed using 2X Maxima SYBR Green/ROX qPCR Master Mix (Thermo Fisher) and gene-specific primers for sopB (CaSopBDETF + CaSopBDETR). rt-qPCRs were monitored and analysed with a Bio-Rad CFX96 Real-Time System and software.
Relative expression was calculated based on the ΔCT values obtained by subtracting the CT value of the house-keeping gene with the gene of interest. gapA, ActB and ACT1 were used as house-keeping genes to normalize Salmonella, mouse immune and C. albicans gene expression, respectively.
Metabolomics analyses
Amino acids were quantified using a Waters Acquity uPLC System with an AccQ-Tag Ultra C18 1.7-μm 2.1 × 100-mm column and a Photodiode Detector Array. Faecal samples were homogenized in methanol (5 μl mg−1 stool) and centrifuged twice at 13,000g for 5 min. Intestinal flushes were vortexed for 1 min and centrifuged twice at 13,000g for 5 min. Amino acids in the supernatant were derivatized using the Waters AccQ-Tag Ultra Amino Acid Derivatization Kit (Waters Corporation) and analysed using the UPLC AAA H-Class Application Kit (Waters Corporation) according to the manufacturer’s instructions. Blanks and standards were run every eight samples. All chemicals and reagents used were mass spectrometry grade.
β-Galactosidase assay
β-Galactosidase assays were performed as previously detailed82. Promoter activity was measured by monitoring β-galactosidase expression from chromosomal transcriptional reporter fusions, as previously described83.
Statistical analyses
Statistical analyses were performed using GraphPad (v10.2.2). All in vitro experiments were conducted at least in triplicate. In all graphs, each symbol represents an independent sample (n), and the individual sample size per experiment has also been indicated in the figure legends. Statistical test used in each experiment has been described in the figure legends.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.