Mice
C57BL/6 mice, maintained under GF conditions, were purchased from Sankyo Laboratories Japan, SLC Japan or CLEA Japan. GF and gnotobiotic mice were bred and maintained within the gnotobiotic facility of Keio University School of Medicine or the JSR-Keio University Medical and Chemical Innovation Center. Il10â/â and Ifngr1â/â mice were purchased from Jackson Laboratories. Myd88â/âTicam1â/â and Rag2â/âIl2rgâ/â mice were purchased from Oriental Bio Service. All mice were maintained under a 12-h lightâdark cycle. A temperature of 20â24â°C and a humidity of 40â60% were used for the housing conditions. All animal experiments were approved by the Keio University Institutional Animal Care and Use Committee.
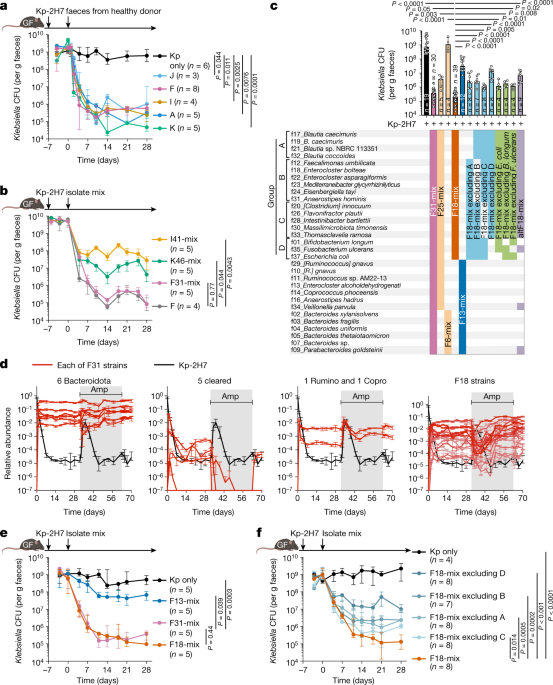
Human faecal samples and isolation of bacterial strains
Human faecal samples were obtained from healthy human donors, patients with ulcerative colitis and patients with Crohnâs disease following the protocol approved by the Institutional Review Board of Keio University School of Medicine (approval numbers 20150075, 20140211, and 20150075). Informed consent was obtained from each individual. Faecal samples were mixed with PBS (containing 20% glycerol) and stored at â80â°C. An aliquot of each sample was diluted with PBS in an anaerobic chamber (80% N2, 10% H2 and 10% CO2; Coy Laboratory Products) and plated onto different agar plates (EG, mGAM, BHK, CM0151, MRS or BL media). After incubating for 2â7 days, colonies with different appearances were transferred to liquid media (EG, mGAM, HK or CM0149), incubated for 24â48âh, mixed with glycerol (final concentration 20% (v/v)), and stored at â80â°C. Bacterial genomic DNA was extracted from the isolated strains using the same protocol as DNA isolation from faecal samples. The 16S rRNA gene locus was amplified by PCR using the KOD plus Neo kit (TOYOBO) according to the manufacturerâs protocol. DNA sequencing was performed by Eurofins. Sequences were aligned using the BLAST program of NCBI and the Ribosomal Database Project (RDP) databases. Primers used for DNA sequencing were as follows: F27 primer: 5â²-AGRGTTTGATYMTGGCTCAG-3â²; R1492 primer: 5â²-TACGGYTACCTTGTTACGACTT-3â². Individual isolates in the culture collection were grouped as âstrainsâ if their 16S rRNA gene sequences shared >98.0% homology.
To prepare the bacterial mixture for inoculation, isolated strains were individually cultured in the appropriate broth at 37â°C for 1â2 days (mGAM broth was used for culturing the F18 strains). Bacterial density was adjusted based on absorbance at 600ânm, and equal volumes of the cultured strains were mixed and centrifuged at 3,000g for 10 min at 4â°C to concentrate fivefold. Thereafter, GF mice were administered 200âμl of the bacterial mixture per mouse (approximately 1â2âÃâ109 CFU of total bacteria) by oral gavage. The bacterial mixture was administered into GF mice (200âμl per mouse, approximately 1â2âÃâ109 CFU of total bacteria) by oral gavage. In Extended Data Fig. 3e, f37_E. coli strain was swapped out for the E. coli Nissle1917 strain (Mutaflor, DSM 6601).
Effect of defined consortia on pathogenic and commensal bacterial strains
To examine the effects of defined consortia on pathogenic bacteria, C57BL/6 GF mice (8â14 weeks of age, housed in separate GF isolators) were inoculated with K. pneumoniae 2H7 (Kp-2H7), carbapenem-resistant K. pneumoniae (CPM+ Kp, ATCC BAA1705), K. aerogenes (strain Ka-11E1217), extended-spectrum-β-lactamase producing E. coli (ESBL+ E. coli, ATCC BAA2777), adherent-invasive E. coli (AIEC, strain LF82, provided by N. Barnich23), P. aeruginosa (ATCC 10145), vancomycin-resistant E. faecium (VRE Ef, ATCC 700221), C. upsaliensis (ATCC BAA1059), or C. difficile (strain 630, ATCC BAA1382) by oral gavage (2âÃâ108âCFU per mouse). Seven days after colonization with pathogenic microbes, the mice were administered 200âμl of isolated bacterial strain mix (total 109 CFU) or 200âμl of human faecal suspension by oral gavage. Faecal samples were collected from mice every three or four days, suspended in PBS (containing 20% glycerol), and cultured on selective media (DHL agar with 30âmgâlâ1 ampicillin and 30âmgâlâ1 spectinomycin for Kp-2H7, CPM+ Kp, Ka-11E12 and P. aeruginosa, MacConkey agar with 1âmgâlâ1 cefotaxime, and VRE-selective agar plates (BD 251832) for VRE). After 24â48âh of incubation, the CFUs were counted. In cases where evaluation was not possible by counting CFUs, bacterial DNA extracted from faeces was evaluated by quantitative real-time PCR (qPCR). To evaluate colonization resistance activity against Kp-2H7, C57BL/6 GF mice were first colonized with either F18-mix or F13-mix and then inoculated with Kp-2H7 on day 7. Faecal samples were collected every three or four days to count Kp-2H7 CFUs. To examine the cumulative effect of the 18 strains on Kp-2H7, C57BL/6 GF mice were inoculated with Kp-2H7 (2âÃâ108âCFU per mouse) by oral gavage, followed by oral administration of each strain of the F18-mix one by one every five days for 95 days. Faecal samples were collected every five days to count the CFUs of Kp-2H7 as well as to quantify the levels of gluconate.
To investigate the influence of F18-mix on commensal strains, 7 strains (Dorea longicatena, Eubacterium rectale, C. scindens, Bacteroides thetaiotaomicron, Bacteroides uniformis, Bifidobacterium adlescentis and Collinsella aerofaciens) were selected from our culture collection. C57BL/6 GF mice were colonized with the 7 commensal strains together with Kp-2H7 (each 2âÃâ108 CFUs). Fourteen days later, mice were administered with F18-mix (total 109 CFUs in 200âµl) by oral gavage. Faecal samples were collected twice per week and subjected to quantification by CFU calculation and qPCR. To investigate the effect of F18-mix in the context of a more complex microbiota, C57BL/6 GF mice were colonized with either 41 strains from donor I, 46 strains from donor K, or a combination of both groups with C. scindens VE202-26 (totaling 88 strains). Subsequently, without any prior antibiotic treatment, these mice were orally inoculated with F18-mix. Faeces were collected for full-length 16S rRNA gene sequencing analysis.
To examine the effects of F18-mix in the context of dysbiotic microbiota, GF B6 or Il10â/â (B6 background) mice were administrated with 200âµl of faecal suspension from patients with ulcerative colitis or Crohnâs disease containing high levels of ESBL+ E. coli or K. pneumoniae. Ten days after IBD microbiota inoculation, mice were treated with 500 μl of 1âgâlâ1 vancomycin by oral gavage. The mice were treated with or without F18-mix four times (at 4, 8, 24 and 48âh following the vancomycin treatment). Faeces were collected and subjected to 16S rRNA gene sequencing or counting CFUs of E. coli or K. pneumoniae.
Dietary supplementation with carbohydrates
Unless otherwise indicated, mice were fed a nutrient-rich diet (CL-2; CLEA Japan), which is high in gluconate. To assess the impact of dietary carbohydrate supplementation, a formula diet (AIN93G; Oriental Yeast Co.) was used, which is high in glucose but lacks gluconate, sorbitol, mannose, xylose, glucarate, galacturonate and xylose. This diet was supplemented with varying percentages of gluconate (0%, 2.5% or 10% of total calories) or with 10% of other carbohydrates, including glucarate, galactitol, sorbitol, cellobiose, glucosamine, xylose, mannose or galacturonate (Supplementary Table 8). The diets were sterilized by γ-irradiation (50âGy).
Bacterial DNA extraction and quantitative real-time PCR
The frozen faecal samples were thawed and 50âμl of each sample was mixed with 350âμl TE10 (10 mM Tris-HCl, 10 mM EDTA) buffer containing RNase A (final concentration 100âμgmlâ1, Invitrogen) and lysozyme (final concentration 3.0âmgâmlâ1, Sigma). The suspension was incubated for 1âh at 37â°C with gentle mixing. Purified achromopeptidase (Wako) was added to a final concentration of 2,000âunitsâmlâ1, and the sample was further incubated for 30 min at 37â°C. Then, sodium dodecyl sulfate (final concentration 1%) and proteinase K (final concentration 1âmgâmlâ1, Nacalai) were added to the suspension and the mixture was incubated for 1âh at 55â°C. Thereafter, purified DNA was obtained from the samples using the Maxwell RSC cultured cell DNA kit, according to the manufacturerâs protocol. For quantifying the amount of bacterial DNA, real-time qPCR was performed using the Thunderbird SYBR qPCR Mix (TOYOBO) and LightCycler 480 (Roche). The primer pairs used in this study are listed in Supplementary Table 9.
Full-length 16S rRNA gene amplicon sequencing
Full-length 16S rRNA amplicon sequencing was performed according to the protocol prepared by PacBio with slight modifications. In brief, the full-length 16S rRNA gene (including hypervariable regions V1 to V9) was amplified using barcoded 27Fmod (5â²-Phos-GCATCNNNNNNNNNNAGRGTTYGATYMTGGCTCAG-3â²) and barcoded 1492R (5â²-Phos-GCATCNNNNNNNNNNRGYTACCTTGTTACGACTT-3â²) primers; âPhosâ indicates a 5â²-phosphate modification, and âNâ represents a unique PacBio barcode sequence for each sample. The PCR conditions were as follows: an initial denaturation at 95â°C for 3âmin, followed by 20 cycles of denaturation at 95â°C for 30âs, annealing at 57â°C for 30âs, and extension at 72â°C for 60âs. The PCR products were purified using AMPure magnetic beads and pooled at equimolar concentrations. The pooled amplicons were further purified with AMPure beads, and 1âμg was used for library preparation. The library was prepared according to the PacBio SMRTbell Prep Kit 3.0 protocol and sequenced on the PacBio Revio system. The HiFi reads were automatically generated using SMRT Link software (version 13.0) with default settings and demultiplexed using lima application in SMRT Tools with HIFI-ASYMMETRIC presets.
Amplicon sequence variants analysis
Full-length 16S rRNA gene amplicon sequence variants (FL16s-ASVs) were inferred from demultiplexed HiFi reads using the DADA2 package (version 1.30.0) in R (version 4.3.3) according to the previously described DADA2 for PacBio workflow53 with slight modifications. The reads were subjected to quality filtering and trimming using the filterAndTrim function with the following parameters: minQ=3, minLen=1300, maxLen=1600, maxN=0, rm.phix=FALSE, maxEE=2. To learn error rates using the learnErrors function without the âdada2:::PacBioErrfunâ option, due to the binned quality value adopted by the Revio system, with a maximum of QV40. FL16s-ASVs were then subjected to a homology search against 16S rRNA gene sequences extracted from publicly available genomes (downloaded from GenBank on 12 September 2023) using BLASTN with a maximum e-value cut-off of 1âÃâ10â10. Top hits were determined by the highest bitscore.
Bacterial whole-genome sequencing
The Illumina MiSeq and PacBio Sequel platforms were used for bacterial whole-genome sequencing. For Illumina sequencing, the library was prepared using the TruSeq DNA PCR-free library prep kit (Illumina), with a target insert size of 550 bp. All the Illumina reads were trimmed and filtered using the FASTX-toolkit (version 0.0.13). For the PacBio sequencing, the library was prepared using the SMRTbell template prep kit 1.0. Sequence data for both types of sequencing were assembled using the hybrid assembler Unicycler. Taxonomic assignment of the genomes was determined by classify_wf of GTDB-tk54 version 2.3.0 with GTDB55 database R214. NCBI taxonomy of fastANI reference genome related to the genome of each strain was retrieved using NCBI-genome-download version 0.3.3 (ncbi-genome-download; https://doi.org/10.5281/zenodo.8192432) and rankedlineage.dmp from NCBI taxonomy database56 (downloaded on 14 September 2023). The genes were predicted using Prokka version 1.14.0 with â–kingdom Bacteria –rnammerâ options, and rnammer version 1.2. The homology search for the predicted genes was performed using diamond57 version 2.0.15 with âblastp –evalue 0.00001 –id 30 –query-cover 60 –ultra-sensitiveâ options, with KEGG (downloaded on 19 April 2022)58, COG (downloaded on 19 May 2021)59, VFDB (downloaded on 10 September 2022)60, and UniRef90 (downloaded on 24 May 2022; https://www.uniprot.org/help/uniref) databases. For homology search against KEGG DB, a database was manually constructed from protein sequences with KEGG Ontology (K number) which were extracted from KEGG non-redundant datasets at the species level. We also added homology search for gluconate metabolism genes in our isolated strains with âblastp –evalue 0.00001 –id 20 –query-cover 60 –ultra-sensitiveâ options. The sequences of gluconate kinase (gntK, MKMCEHOJ_02531) and gluconate transporters (MKMCEHOJ_02530 and MKMCEHOJ_02505) from f37_E. coli strain, and gluconate dehydratase (gad, EAOGLLOI_00767), gluconate transporters (EAOGLLOI_00766 and EAOGLLOI_00912), 2-dehydro-3-deoxygluconokinase (kdgK, EAOGLLOI_00768), and 2-dehydro-3-deoxyphosphogluconate aldolase (eda, EAOGLLOI_00769) from f17_Blautia caecimuris strain were used as reference sequences.
Ex vivo caecal suspension culture
Caecal contents from GF or F31-mix, F18-mix and F13-mix colonized mice were anaerobically resuspended in water at a concentration of 100âmgâmlâ1. Caecal contents were either filtered through a 0.22-μm filter (Millex Millipore) after centrifuging at 10,000g for 5âmin, heat-killed at 105â°C for 30 min, or left untreated. Thereafter, a diluted overnight culture of Kp-2H7 (103 CFU in 10âμl) was added to 200âμl of each caecal suspension. After incubating at 37â°C for 48âh under aerobic or anaerobic conditions, samples were serially diluted and plated on a selection agar plate (DHL with 30âmgâlâ1 ampicillin and 30âmgâlâ1 spectinomycin) for counting Kp-2H7 CFU.
Bacterial growth monitoring
The wild-type, ÎgntK or ÎgntR Kp-2H7 strain was cultured in M9 minimal medium for 24âh at 37â°C. Afterward, the culture was diluted 100-fold with sterile water. A 10-μl aliquot of the diluted culture was inoculated into 200âμl M9 medium supplemented with individual carbohydrates (final concentration of 2 mM) as the sole carbon source, or with a mock control. To examine the effect of metabolites on Kp-2H7 growth, 10âμl of wild-type Kp-2H7 culture dilutions were inoculated into 200âμl M9 medium containing varying concentrations of 4-HBA (100, 10, 1 or 0.1âmM), cholic acid (500, 100, 20 or 4âμM), and acetate or butyrate (100, 25, 6.25, 1.56 or 0.39âmM). The pH of acetate and butyrate was adjusted to either 5.0 or 7.0. Bacterial growth was monitored by measuring absorbance at 600 nm every 30âmin using a microplate reader (Sunrise Thermo (Tecan) for anaerobic conditions and Infinite 200 PRO (Tecan) for aerobic conditions) at 37â°C with 100âs shaking before each time point.
Transcriptome analysis of epithelial cells
Total RNA was isolated from colonic epithelial cells using NucleoSpin RNA (Macherey-Nagel), according to the manufacturerâs instructions. Libraries for RNA sequencing were prepared using TruSeq Stranded mRNA Library Prep (Illumina), according to the manufacturerâs instructions. The libraries were sequenced using NovaSeq 6000 (Illumina) with the mode of 150-bp paired-end. The sequenced paired-end reads were quality-controlled using Trimmomatic61 version 0.39 with â2:30:10 LEADING:3 TRAILING:20 SLIDINGWINDOW:4:15 MINLEN:5â options and FASTX-Toolkit version 0.0.13 (https://github.com/agordon/fastx_toolkit) with â-q 20 -p 80â options. Unpaired reads and reads mapped to the PhiX reference genome using minimap262 version 2.17-r941 were excluded from further analyses. The remaining quality-controlled reads were mapped to the mouse reference genome (mm10) using STAR63 version 2.7.2b. The mapped reads were counted for each gene using featureCounts64 version 1.5.2 with â-t exon -p -B -Q 1â options. the transcripts per million (TPM) values of each gene in each sample were calculated. The differential expression analysis was performed using DESeq265 version 1.28.1, and the P values were corrected by the BenjaminiâHochberg method to maintain the false discovery rate (FDR) below 5%.
Transcriptome analysis of Kp-2H7
To examine the transcriptome landscape of Kp-2H7 in vivo, GF mice were monocolonized with Kp-2H7, followed by oral administration of F18-mix or vehicle control. Two days after F18-mix administration, faecal samples were collected, and total RNA was extracted. For the in vitro examination of the transcriptomes of Kp-2H7 strains, wild-type, ÎgntK, and ÎgntR Kp-2H7 were cultured at 37â°C in M9 minimal medium supplemented with either glucose or gluconate. Bacteria were collected during the early log phase (absorbance at 600ânmâ=â0.35), and total RNA was extracted. Isolation of total RNA from in vivo faecal samples or in vitro culture samples was conducted using the NucleoSpin RNA kit (Macherey-Nagel), according to the manufacturerâs instructions. Libraries for RNA sequencing were prepared using TruSeq Stranded mRNA Library Prep (Illumina) and sequenced using HiSeq X (Illumina) with the mode of 150-bp paired-end. To analyse the in vivo transcriptome profiles of Kp-2H7 in the presence or absence of F18-mix, a reference genome was created by concatenating the genome sequence of Kp-2H7 with the genome sequences of the F18-mix. The sequenced paired-end reads were quality-controlled using Trimmomatic61 version 0.39 with â2:30:10 LEADING:3 TRAILING:20 SLIDINGWINDOW:4:15 MINLEN:5â options and FASTX-Toolkit version 0.0.13. Unpaired reads and reads mapped to the mouse (mm10) or PhiX reference genome using minimap262 version 2.17-r941 (in vivo) or 2.24-r1122 (in vitro) were excluded from further analyses. The quality-controlled reads were mapped to the concatenated or Kp-2H7 reference genome using bowtie266 version 2.3.4.1. (in vivo) or 2.4.4 (in vitro). For in vivo mice faecal samples, the read counts for each Kp-2H7 gene were obtained by counting uniquely mapped reads and then distributing and summing multi-hit read counts based on the number of uniquely mapped reads. For in vitro culture samples, the read counts for each Kp-2H7 gene were obtained using featureCounts64 version 2.0.1 with â-t CDS -p -B -Q 10â options. The differential expression analysis was performed using DESeq265 version 1.28.1 (in vivo) or 1.30.1 (in vitro) with âfitType = localâ option and BenjaminiâHochberg correction method to maintain the FDR below 5%. The heat map was obtained from the variance-stabilizing transformations values obtained from the DESeq2 output.
For real-time qPCR analysis, cDNA was synthesized using ReverTra Ace qPCR RT Master Mix (TOYOBO), and qPCR was performed using Thunderbird SYBR qPCR Mix (TOYOBO) on a LightCycler 480 (Roche).
Construction of transposon mutant library
A transposon insertion library of Kp-2H7 was constructed using the EZ-Tn5TM <KAN-2> Tnp Transposome kit (Lucigen). In brief, 80âμl (109 CFU) of Kp-2H7 suspension was mixed with 0.5âμl of EZ-Tn5TM <KAN-2>, transferred to a 1-mm gap width electroporation cuvette, and subjected to electroporation using ELEPO21 (Nepa Gene Co.) with the following parameters: poring pulse; voltage: 1,800âV, pulse length: 5.0âms, pulse interval: 50âms, number of pulses: 1, and polarity: +, and transfer pulse; voltage: 150âV, pulse length: 50âms, pulse interval: 50âms, number of pulses: 5, and polarity: ±. Transformed Kp-2H7 cells were incubated in 1âml LB broth for 3âh at 37â°C, and then selected on LB agar plates containing kanamycin (90âmgâlâ1) at 37â°C. Thereafter, approximately 8âÃâ105 transposon mutant colonies were collected and stored at â80â°C in LB containing 20% glycerol.
Transposon sequencing
GF mice were colonized with the pool of 8âÃâ105 Kp-2H7 transposon mutants. Faecal samples were collected on days 0, 4, 10 and 28 following colonization, suspended in PBS (50âmgâmlâ1) containing 20% glycerol, and cultured overnight at 37â°C on LB agar plates containing kanamycin (90âmgâlâ1). Kp-2H7 mutant colonies were scraped together and DNA was extracted by the method described above. Transposon sequencing was carried out according to the method described by Kazi et al.67. In brief, genomic DNA was fragmented via sonication. Then, a poly-C tail was added to the 3â² end of the DNA fragment by terminal deoxynucleotidyl transferase. The transposon junctions were amplified using a biotinylated primer, which was then enriched using streptavidin beads. By performing a second nested PCR, a single barcode was added to each sample. The libraries were sequenced using HiSeq 2500 (Illumina) with the mode of 50-bp single-end. The first 24 bases of each sequenced read were trimmed to exclude primer and mosaic end sequences. The trimmed reads were quality-controlled using Trimmomatic61 version 0.39 and FASTX-Toolkit version 0.0.13. The remaining reads were mapped to the PhiX reference genome (mm10) using minimap2 version 2.17-r941 to exclude those that align with the PhiX genome. Then, the analysis-ready reads were mapped to the Kp-2H7 genome using bowtie2 version 2.4.266. The mapped reads were counted for each gene using featureCounts64 version 1.5.2 with â-t CDS -p -B -Q 1â options, and the TPM of each gene was calculated as the relative abundance of a gene mutant in a sample by assuming that each transposon mutant has a single insertion. The differential abundance mutants were detected by Welchâs t-test for log-scaled TPM with BenjaminiâHochberg correction method to maintain the FDR below 5%.
Generation of Kp-2H7 mutants
The Kp-2H7 deletion mutants were generated as shown in Supplementary Fig. 3 using the Quick and Easy E. coli Gene Deletion Kit (Gene Bridges) according to the manufacturerâs protocol. In brief, Kp-2H7 cells were transformed with the pRED/ET plasmid harbouring the tetracycline-resistant gene by electroporation. Bacteria with pRED/ET were selected on LB plates containing tetracycline (30âmgâlâ1) at 30â°C. Thereafter, these cells were incubated in LB broth with appropriate antibiotics at 30â°C until absorbance at 600 nm reached 0.2, followed by an additional 1âh of incubation with 0.3% l-arabinose at 37â°C to induce the expression of the recombinant proteins. These cells were used to prepare electrocompetent cells and were transformed with the linear DNA fragment (the FRT-PGK-gb2-neo-FRT cassette)-flanked homology arms. The functional cassettes were generated by PCR, according to the manufacturerâs protocol. The primers with homology arms are listed in Supplementary Table 9. The electroporated cells were incubated in 1âml LB broth for 3âh at 37â°C. Gene deletion strains were selected on LB agar plates with kanamycin (90âmgâlâ1) after overnight growth at 37â°C. The double-knockout strains were generated by removing the kanamycin selection marker through electroporation of the FLP expression plasmid (707-FLPe) and repeating the above-mentioned protocol. The deletions were confirmed by DNA sequencing.
Isolation of lymphocytes and flow cytometry
Lymphocytes were collected from the large intestines and analysed according to previously described protocols17,68. In brief, the intestines were dissected longitudinally and washed with PBS to remove all luminal contents. All samples were incubated in 15âml Hanksâ balanced salt solution (HBSS) containing 5 mM EDTA for 20 min at 37â°C in a shaking water bath to remove epithelial cells. Thereafter, after removal of any remaining epithelial cells, muscular layers and fat tissues using forceps, the samples were cut into small pieces and incubated in 10âml RPMI1640 containing 4% foetal bovine serum (FBS), 0.5âmgâmlâ1 collagenase D (Roche Diagnostics), 0.5âmgâmlâ1 dispase II (Roche Diagnostics), and 40âμgâmlâ1 DNase I (Roche Diagnostics) for 50âmin at 37â°C in a shaking water bath. Thereafter, the resultant digested tissues were washed with 10âml HBSS containing 5 mM EDTA, resuspended in 5âml of 40% Percoll (GE Healthcare), and underlaid with 2.5âml of 80% Percoll in a 15-ml Falcon tube. Percoll gradient separation was performed by centrifugation at 850g for 25 min at 25â°C. Lymphocytes were collected from the interface of the Percoll gradient and washed with RPMI1640 containing 10% FBS, and then stimulated with 50ângâmlâ1 PMA and 750ângâmlâ1 ionomycin (both from Sigma) in the presence of Golgistop (BD Biosciences) at 37â°C for 4âh. After labelling of the dead cells with Ghost Dye Red 780 Viability Dye (Cell Signaling Technology), the cells were permeabilized and stained with anti-CD3e (BUV395; BD Biosciences), anti-CD4 (BUV737; BD Biosciences), anti-TCRβ (BV421; Biolegend) and anti-IFNγ (FITC; Biolegend) at 1:1,000 dilution using the Foxp3/Transcription Factor Staining Buffer Kit (Tonbo Biosciences), according to the manufacturerâs instructions. All data were collected on a BD LSRFortessa (BD Biosciences) and analysed using Flowjo software (TreeStar). CD4+ T cells were defined as a CD4+TCRβ+CD3e+ subset within the live lymphocyte gate.
Measurement of lipocalin-2 and calprotectin
The faecal pellets from Il10â/â mice were vortexed, suspended in PBS (5% w/v) with Complete Protease Inhibitor Cocktail (1 tablet dissolved in 50âml PBS; Roche) and centrifuged, and supernatants were collected. The concentration of lipocalin-2 and calprotectin in faecal supernatants was measured by ELISA (Mouse Lipocalin-2 Matched Antibody Pair Kit; Abcam, Mouse S100A8/S100A9 Heterodimer DuoSet; R&D), according to the manufacturerâs protocol.
Histological analysis
Colon tissue samples were dissected longitudinally and swiss-rolled, fixed with 4% paraformaldehyde, embedded in paraffin, sliced to 5-μm sections and stained with hematoxylin and eosin. The degrees of colitis were graded by the mouse colitis histology index69. The histological slides were evaluated blind by two investigators.
Non-targeted metabolomics analysis
C57BL/6 GF mice were monocolonized with Kp-2H7, followed by oral administration of bacterial mix. Caecal contents were collected on day 28 after administration of isolated bacterial mix and stored at â80â°C until use. Frozen caecal contents were homogenized by shaking with metal corn using a multi beads shocker as previously described70. Then, the samples were suspended in 400âμl of methanol per 100âmg caecal contents, and a 40âμl aliquot was subjected to the single layer extraction and untargeted LCâQTOF/MS analysis70. SCFAs were simultaneously extracted and derivatized from 20âμl of the suspension by using pentafluorobenzyl bromide alkylation reagent (Thermo Fischer Scientific), and analysed by gas chromatographyâmass spectrometry (GCâMS) as previously described71. Water-soluble metabolites were extracted by first mixing 4âμl of the suspension, 196âμl of methanol, 200âμl of chloroform, 70âμl of water, and 10âμl of internal standards mix (100âμM cycloleucine, 500âμM citric acid-d4, and 1.0âmM ornithine-d7 (Cambridge Isotope Laboratories)). After vortexing for 1âmin and centrifugation at 15,000g for 5 min at 4â°C, 100âμl of supernatant was evaporated to dryness. The dried samples were derivatized via methoxyamination, trimethylsilylation, or tert-butyldimethylsilylation, and then analysed by GCâMS/MS using Smart Metabolite Database (Shimadzu) or GCâMS operated in selected ion monitoring mode, as described previously72. Bile acids were extracted from 4âμl of the suspension mixed with deuterium-labelled internal standard mix (1.0 μM cholic acid-d4, 1.0 μM lithocholic acid-d4, 1.0 μM deoxycholic acid-d4, 1.0 μM taurocholic acid-d4, and 1.0 μM glycocholic acid-d4 (Cayman Chemical)) using the Monospin C18 column (GL science). The column was washed with 300âμl water (Ã2) and 300âμl of hexane (Ã1). Bile acids were eluted with 100âμl methanol, then subjected to LCâMS/MS analysis using an UPLC I class (Waters) with a linear ion-trap quadrupole mass spectrometer (QTRAP 6500; AB SCIEX) equipped with an Acquity UPLC BEH C18 column (50 mm, 2.1 mm, and 1.7 μm; Waters). Samples were analysed with a mobile phase consisting of water:methanol:acetonitrile (14:3:3 (vol:vol:vol)) and acetonitrile, both containing 5 mM ammonium acetate, for 4 min, which was changed to 40:60 after 12 min, to 5:95 after 2 min, and then held for 2 min; with flow rates of 300âμlâminâ1. Bile acids were detected by multiple-reaction monitoring in negative mode. Ions of [M-H]â, taurine (m/zâ=â124), and glycine (m/zâ=â74), generated from the precursor ion, were monitored as product ions for non-conjugated, taurine-conjugated, and glycine-conjugated bile acids, respectively. MS/MS settings were as follows: ion source, turbo spray; curtain gas, 30âpsi; collision gas, 9âpsi; ion spray voltage, â4,500âV; source temperature, 600â°C; ion source gas 1, 50âpsi; and ion source gas 2, 60âpsi.
Measurement of carbohydrate levels
To evaluate bacterial gluconate utilization in vitro, isolated strains were cultured in mGAM broth or RCM containing 300âμM gluconate for 48âh at 37â°C under anaerobic conditions. Supernatant of each culture broth was collected, and the concentration of gluconate was measured by the ExionLC AD and SCIEX Triple Quad 6500+ LCâMS/MS system. To evaluate carbohydrate levels in faeces or intestinal contents, each sample was suspended in water (50âmgâmlâ1), and the carbohydrate levels in the supernatant were measured by LCâMS/MS. The measurement conditions for gluconate, glucuronate, and galacturonate were as follows: chromatographic separation was performed using the Intrada Organic Acid column, 150âÃâ2âmm (Imtakt); column temperature was 40â°C; and the volume of each injection was 5âμl. The mobile phase comprising A (acetonitrile/water/formic acid, 10/90/0.1) and B (acetonitrile/100mM ammonium formate, 10/90) was used under gradient conditions: 0â1.5âmin, A 100%, B 0%; 1.6â7âmin, A 70%, B 30%; 10â13âmin, A 0%, B 100%; and 13.1â18âmin, A 100%, B 0%); and the flow rate was 0.2âmlâminâ1. Detailed MS conditions were as follows: curtain gas, 30âpsi; collision gas, 6; ion spray voltage, â4,500âV; temperature, 550â°C; ion source gas 1, 50âpsi; and ion source gas 2, 60âpsi. The retention time and multiple-reaction monitoring transitions are listed in Supplementary Table 10. The measurement conditions for other carbohydrates were as follows: chromatographic separation was performed using the UK-Amino column (UKA26), 250âÃâ2âmm, (Imtakt); column temperature was 65â°C and the volume of each injection was 2âμl. The mobile phase comprising A (5âmM ammonium acetate, 0.05% formic acid) and B (acetonitrile) was used under gradient conditions: 0â10âmin, A 5%, B 95%; 35âmin, A 15%, B 85%; 50âmin, A 40%, B 60%; 50.1â55âmin, A 80%, B 20%; 55.1â60âmin, and A 5%, B 95%); and the flow rate was 0.25âmlâminâ1. Detailed MS conditions were as follows: curtain gas, 25âpsi; collision gas, 9; ion spray voltage, â4,500âV in negative mode and 5,500âV in positive mode; temperature, 250â°C, ion source gas 1, 50âpsi; and ion source gas 2, 70âpsi. Multiple-reaction monitoring parameters are listed in Supplementary Table 10. Data were obtained using Analyst software version 1.7.1 and analysed using SCIEX OS-MQ software version 2.1.0.55343.
Metagenomic analysis of IBD cohorts
To explore established and novel microbial taxa possessing gluconate operon genes, gene catalogues were acquired from two cohorts with IBD aetiology: the paediatric PROTECT and adult HMP2 cohorts, comprising 240 and 1,638 longitudinal metagenomic samples from 94 and 91 individuals, respectively. MSPs were constructed via the co-abundant gene binning (MSPminer73), followed by quality assessment (CheckM74), as described by Schirmer et al.3 (PROTECT) and Kenny et al.75 (HMP2). A targeted screening of these bins with DIAMOND BLASTP version 0.9.1476 was conducted to identify putative gluconate transport and metabolism genes, retaining hits with an e-value <0.01 and sequence identity â¥60%. MSPs were categorized based on the combinations of gluconate-related genes detected. A differential abundance analysis was performed on TPM-normalized and centred log-ratio-transformed MSP counts to control for sequencing depth, gene length, and compositional biases. Statistical significance was ascertained through a non-parametric, two-sided MannâWhitney U test with BenjaminiâHochberg correction. Effect sizes (r), calculated as the test statistic divided by the square root of the sample size, along with bootstrapped confidence intervals, were computed to account for unbalanced group sizes, offering insights into the robustness and directionality of the observed effects.
For PROTECT, comparative analyses were iteratively repeated with varying seed values for random sample selection from longitudinal data pools of mild (nâ=â64), moderate/severe (nâ=â57), and non-IBD samples (nâ=â119). Within HMP2, inclusion was limited to cross-sectional samples accompanied by calprotectin data. In response to the attenuated disease signal observed in the study cohort77, a targeted inflammation-specific selection approach was utilized. For IBD cases the sample with maximal calprotectin value per patient was included (Crohnâs disease, nâ=â41; ulcerative colitis, nâ=â26). Conversely, for the non-IBD control group, the sample with the minimal calprotectin value per patient was chosen (nâ=â24). Statistical analyses were conducted using R software version 4.2.1 (Ubuntu 20.04.5 LTS).
Untargeted stool metabolomics and gluconate intensity estimation
Untargeted stool metabolomics of faecal samples from the PROTECT cohort was performed using LCâMS in negative mode, and calprotectin was measured by ELISA. In brief, hydrophilic interaction liquid chromatography (HILIC) analyses of water-soluble metabolites in the negative ionization mode were conducted using Shimadzu Nexera X2 U-HPLC (Shimadzu) coupled to a Q Exactive Plus mass spectrometer (Thermo Fisher Scientific). Metabolites were extracted from plasma or stool (30âμl) using 120âμl of 80% methanol containing inosine-15N4, thymine-d4, and glycocholate-d4 internal standards (Cambridge Isotope Laboratories). The samples were centrifuged (10 min, 9,000g, 4â°C), and the supernatants were injected directly onto a 150âà 2.0 mm Luna NH2 column (Phenomenex). All masses detected in HILIC negative mode were matched via adduct subtraction and molecular formula match to compounds downloaded from the Human Metabolome Database (HMDB) on 10 October 2022. The measured m/z values were adjusted for [M-H]- adducts, and molecular formulae matching to within 5 ppm were selected as candidate identifiers. In cases where multiple molecular formulae matched the adduct-adjusted mass (as a result of multiple potential adducts), the one with a minimal ppm difference was selected. Out of 4,461 detected features (m/z, retention time pairs), a single feature 195.0512 m/z at 4.34 min resolved to the formula C6H12O7 (delta ppm = 0.89), related to a group of 5 compounds with canonical structure O=C(O)C(O)C(O)C(O)C(O)CO, which includes l-gluconic acid (HMDB0000625). The metabolic feature was subsequently validated with a reference standard (Sigma Aldrich, S2054) via retention time and MS/MS match against iHMP pooled stool samples, and aligned via global m/z and retention time matching with the PROTECT stool samples using Eclipse78. This led to annotating HNs_QI1923 (HILIC-neg 195.0512 m/z at 4.34 min) from PROTECT and QI11027 (HILIC-neg 195.0512 m/z at 4.48 min) from HMP2.
Statistical analyses
Statistical analyses were performed using GraphPad Prism software. KruskalâWallis test and the FDR method of Benjamini and Hochberg were used for multiple comparisons during CFU comparisons. MannâWhitney U test with Welchâs correction was used for comparisons between the two groups. Spearmanâs rank correlation was used to investigate the correlation between the relative abundance of Kp-2H7 and isolated strains.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.