Study oversight
Clinical oversight of the vaccine therapy was undertaken at University Medical Centre, Heidelberg. The patient was treated with a personalized peptide vaccine within the scope of an individual healing attempt (statement WD 9–3000–083/23 of the German Parliament, guidelines 2001/20/EG and 2005/28/EG, Declaration of Helsinki of the World Medical Association (Article 37)); approval by the Institutional Review Board and ethics committees is not required. Informed consent for the vaccine therapy was taken in accordance with local policies. Informed consent for genetic and immune research studies was obtained in accordance with protocols approved by the University Medical Centre Heidelberg Institutional Review Board. Written, informed consent to transfer and perform analyses at the Francis Crick Institute and associated institutions was also provided.
DNA sample extraction and sequencing
Fresh frozen and FFPE samples
The methods for DNA extraction and sequencing for fresh frozen and formalin-fixed paraffin-embedded (FFPE) samples are summarized in the TRACERx manuscripts3,33,38. For fresh frozen recurrence/progression samples, paired germline DNA was re-sequenced in the same run, using germline DNA from aliquots extracted at initial germline blood collection. No further germline sequencing was performed for FFPE samples.
WES bioinformatics pipeline
The bioinformatics pipeline, including quality control checks, used for WES data analysis is summarized in the TRACERx manuscripts3,33,38. VarDict (v.2016.11.21) was used to call the VAF of the EGFR ex19del, as it has been shown to have improved estimates of indel allele frequencies47.
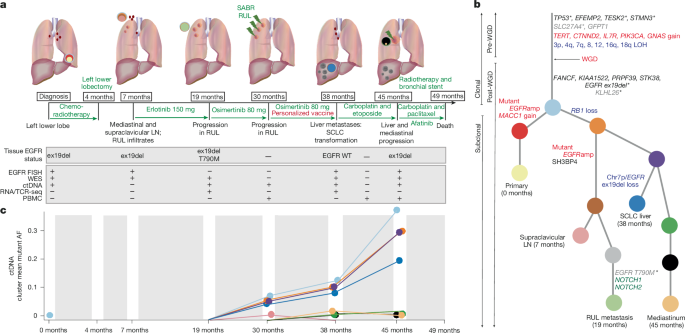
Phylogenetic trees
CONIPHER (COrrecting Noise In PHylogenetic Evaluation and Reconstruction) was used to construct the phylogenetic tree3,48. The tree was manually reviewed/selected, and orthogonal checks were performed (Supplementary Note).
Timing mutations relative to WGD
Strict criteria were used to define a mutation as pre-WGD in a simulated ‘single biopsy’ analysis. Using data from a single region, we inferred whether a variant’s copy number status tracked the major or minor copy number allele. For example, with LOH (that is, minor copy number is 0), the presence of a variant means it must track with the major allele. Where the variant copy number is larger than the minor copy number, it too must track the major allele. Where there is only one major and/or one minor copy of the allele, we cannot infer whether the variant occurred pre- or post-WGD and we categorized these timings as ‘unclear’. Where the variant copy number is less than or equal to the minor copy number, we assume it is tracked with the minor allele. To minimize false categorization of variants as pre-WGD, we performed a proportion test using the mutation’s VAF and used the estimated 95% lower limit VAF to calculate the minimum copy number state for the variant, and used this to define the timing of the variant relative to WGD. Similarly, to minimize falsely categorizing variants as post-WGD, we used the estimated 95% upper limit VAF to calculate the maximum copy number state for the variant and inferred the timing. If the classification of the variant differed when using the upper limit and lower limits, the timing was then defined as ‘unclear’. Where there are two WGD events in a single region, this method times the variant relative to the first WGD event. Thus, when describing variants as ‘pre’ or ‘post’ WGD, we refer to the first WGD event.
For the driver mutation analysis, we leveraged evidence from all regions in the tumour as well as using the maximum copy number state for the variant, calculating from the 95% upper confidence interval from the VAF proportion test to avoid falsely categorizing an event as occurring post-WGD. Where there is subclonal WGD, the presence of a variant in a non-WGD region suggests that the event must have occurred pre-WGD.
HLA LOH prediction
HLA LOH prediction for the sequenced regions was performed using LOHHLA27.
Peptide vaccine design and manufacture
The vaccine was manufactured by the GMP & T Cell Therapy core facility (German Cancer Research Centre, DKFZ, Heidelberg, Germany) in accordance with facility standard operating procedures, using variant data from the sequenced supraclavicular LN and the RUL lesion (available sequencing at time of manufacture). netMHCpan 4.0 (ref. 49) was used to predict the affinity of the peptides. Priority was given to variants that seemed clonal at that time/were present in both samples, and that had a high predicted affinity (less than 1,000 nM, and ideally less than 500 nM), resulting in 14 candidate targets (Extended Data Table 1). The exceptions to these criteria are the TP53 p.P118L (lower affinity) and EGFR p.T790M (single sample), which were included because of clinical interest; and GFPT1 p.L598V (lower affinity), which was found at high VAFs in both the supraclavicular LN and RUL lobe. Briefly, for the manufacturing, solid phase synthesis using Fmoc chemistry was applied in a fully automated multiple synthesizer (Syro II, MultiSynTech). Synthesis was carried out on preloaded Wang-resins with 2-(1H-Benzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HBTU) as a coupling agent. More than 5,000 peptides have been manufactured at this facility for research purposes. Quality control checks are in place to safeguard against contamination and ensure correctness of the sequence. The 14 candidate peptides (24–29 amino acids in length) were dissolved in water with 10% DMSO for infusion, and four peptides were found to be insoluble (GFPT1 p.L580V, KLHL26 p.R190W, SLC27A4 p.T329M, EGFR p.T790M). The remaining ten long peptides were used for injection. Each vaccine contained 60 µg in 60 µl per peptide (10 peptides at 600 µl total), and mixed with 600 µl of Montanide ISA 51 to formulate the vaccine. Pooled peptides were injected intradermally.
ctDNA analyses
Patient-specific anchored-multiplex PCR enrichment panels were generated using 467 autosomal somatic mutations detected from the tissue WES output20. Additionally, mutations in genes associated with resistance to EGFR TKI therapy were also explored: these include mutations in PIK3CA, KRAS, NRAS, BRAF and EGFR (Supplementary Table 2). Libraries were prepared according to the ArcherDX LiquidPlex ctDNA protocol for Illumina with the following modifications: the first PCR was performed using these cycling conditions: 95 °C for 3 min, 11 cycles: 95 °C for 30 s and 65 °C for 15 min, followed by 72 °C for 3 min and a hold at 4 °C. The second PCR was performed using these cycling conditions: initial denaturation at 95 °C for 3 min, 15 cycles: 95 °C for 30 s and 65 °C for 15 min, followed by 72 °C for 3 min and a hold at 4 °C. Libraries were sequenced on an Illumina NextSeq sequencer to approximately 50 million read pairs per sample and the resulting FASTQs were analysed using the Archer Analysis circulating free DNA variant calling pipeline20. Copy number aberrations associated with resistance were explored from low pass whole genome sequencing of the circulating free DNA using ichorCNA (v.0.1.0)50.
RNA-seq sample sequencing and bioinformatics pipeline
The extraction and sequencing pipelines are summarized in a previous TRACERx manuscript34. Danaher gene signatures29 and CIBERSORTx30 were used to deconvolute the immune microenvironment.
FISH
FISH was carried out using the Vysis EGFR/CEP7 FISH Probe set (Abbott Molecular) in combination with the Histology FISH Accessory Kit (Agilent Technologies). Freshly cut 4-µM pathology sections were xylene-dewaxed, followed by serial rehydration into FISH buffer. Sections were incubated at 98 °C for 10 min in hybridization pre-treatment solution followed by on-section pepsin digestion for 10 min at 37 °C. After serial dehydration, FISH probes were applied to the section, sealed with rubber cement glue and co-denatured at 71 °C for 5 min. Probe/tissue annealing for 16 h was followed by a 65 °C stringent wash for 10 min. Sections were dehydrated, antifade mounting media containing DAPI (Vectashield) was applied and then sections were visualized using a Zeiss Observer Z1 microscope.
Immune analyses
Tissue culture
Blood samples were collected in Vacutainer EDTA blood collection tubes (BD) and PBMCs isolated within 24 h of apheresis by density gradient centrifugation (750g for 10 min) on Ficoll Paque Plus (GE Healthcare). The interface was washed twice with complete RPMI-1640, and cells were resuspended in 90% FBS with 10% DMSO (Sigma-Aldrich) and cryopreserved in liquid nitrogen.
MANAFEST assay
PBMCs were thawed, washed and seeded at 200,000 cells per well in a 96-well plate, in duplicate, in TexMACS media (Miltenyi) containing 5% human AB serum, penicillin-streptomycin and amphotericin B (all from Sigma-Aldrich), and 10 ng ml−1 human interleukin (IL)-15 plus 50 ng ml−1 human IL-21 (all cytokines from BioLegend), with IL-2 (Proleukin, Clinigen) added on day 1 at a final concentration of 40 IU ml−1. Cells were maintained in culture with regular feeding or passage as required, every 2–3 days, in 5% CO2 at 37 °C for a total of 11 days. 24–29-mer neopeptides were synthesized by a manufacturing process that achieves purity of 95%, in which all peptides showed one major peak at the expected molecular weight (Pepscan/Biosynth). Lyophilized peptides were reconstituted in ultra-pure DMSO (Sigma-Aldrich) and added on day 0 at a final concentration of 1 µg ml−1. A cocktail of 8–12-mer viral peptides derived from human CMV, EBV and flu was used as a positive control (Peptivator, Miltenyibiotec) and added at day 0 at a final concentration of 1 µg ml−1 for each peptide. Cell pellets were collected on day 11 and lysate stored in RLT Buffer at −80 °C before RNA extraction (RNAeasy Mini kit, Qiagen). The number of detectable TCRs at baseline was set to 1 from 0 for fold change visualization for which no response was detected. Ex vivo T cell receptor sequencing (TCR-seq) repertoires were isolated from thawed PBMCs cultured overnight without cytokine stimulation.
TCR-seq
TCR alpha and beta sequencing was performed on RNA extracted from MANAFEST assay PBMC cultures and bulk RNA acquired from the RUL and SCLC-transformed liver metastasis, using a quantitative experimental and computational TCR-seq pipeline described recently51,52. This protocol incorporates a unique molecular identifier attached to each complementary DNA TCR molecule that enables correction for PCR and sequencing errors. The suite of tools used for TCR identification, error correction and CDR3 extraction are freely available at https://github.com/innate2adaptive/Decombinator.
TCR-seq analysis
The 3,000 most abundant unique beta chain CDR3s from each sample were selected for analysis as previously described51. Where multiple clones showed equal abundance at rank 3,000, the count value closest to 3,000 was used as a cut-off. Samples were analysed using backend code from the MANAFEST23 webtool (https://sourceforge.net/projects/manafest/; http://www.stat-apps.onc.jhmi.edu/FEST). Neopeptide-stimulated samples were analysed relative to cytokine alone control from the matched time point. Clones were classified as significantly enriched in a given condition if they exclusively showed an odd’s ratio > 10 and Q < 0.01 by false discovery rate-corrected Fisher’s exact test compared with the no peptide (cytokine only) control condition. Only clones present at 500 or more copies in the test condition were considered for analysis unless otherwise specified, with or without being detected in the control condition. Ex vivo samples yielded fewer than 3,000 unique TCR sequences and were not used for MANAFEST analysis. For visualization and calculation of fold change in the number of detected clones, a 0 value is ascribed a value of 1. Analysis was conducted in R using the dplyr (v.1.1.4), immunarch (v.0.9.1), data.table (v.1.14.8), RColorBrewer (v.1.1-3), viridis (v.0.6.5) and ggplot2 (v.3.5.1) packages.
TCR clustering through Gliph2
To identify groups of TCRs that shared similar sequence structure to the clones that were significantly expanded in PBMC samples from the MANAFEST assay, we clustered together the top 3,000 CDR3B sequences from each time point (months 30, 40, 45), from each condition (Cytokine, CEF, ex19del, T790m), using the Gliph2 clustering algorithm. Gliph2 generates output scores per cluster by quantifying clonal expansion and estimating the likelihood that those sequences will cluster together. More significant clusters with more unique TCR sequences are located towards the centre of the network plot, whereas clusters with weaker connections are located further out. To verify that the expanded sequences were driven by peptide-specific stimulation, we allocated TCR clusters to a condition using a 50% threshold to ensure each cluster was included only once in the analysis and we maximized all available data. We then compared cluster importance scores of cytokine culture alone with other conditions (CEF, ex19del and T790M). For Gliph2 TCR clustering, the top 3,000 CDR3B sequences with matching TRBV genes and a count of 3 or greater from the PBMC samples were clustered together using the ‘gliph2’ function from the turboGliph package53 (v.0.99.2). All productive CDR3B sequences with a corresponding V gene were included. Clusters were assigned to a condition on the basis of a count proportion threshold of more than 50%, so each cluster would be represented only once in the analysis. The cluster importance score is the −log10-transformed value of the ‘total.score’ metric from the Gliph2 output.
Fluorospot
PBMCs were thawed, washed and seeded at 5 × 106 cells per well of a 24-well plate in TexMACS media (Miltenyi) containing 5% human AB serum (Sigma-Aldrich), penicillin-streptomycin (Sigma-Aldrich) and 10 ng ml−1 human IL-15 plus 50 ng ml−1 human IL-21 (both from BioLegend), with IL-2 added on day 2 at a final concentration 40 IU ml−1. A cocktail of all ten vaccine and four non-vaccine long neopeptides was added at a final concentration of 1 µg ml−1 on day 1. Cells were maintained in culture with regular feeding or passage as required, every 2–3 days, in 5% CO2 at 37 °C for a total of 11 days, before washing and re-plating overnight in media deprived of cytokine. Rested cells were plated for Fluorospot analysis at 150,000 cells per well and re-stimulated for 24 h with 1 µg ml−1 peptide, 2 µg ml−1 phytohaemagglutinin (Sigma-Aldrich) or anti-CD3 (1 µg ml−1), plus anti-CD28 (20 µg ml−1) antibodies supplied in the Fluorospot kit for Human GZMB and IFNG used as per the manufacturer’s instructions (MabTech). Plates were protected from light until being read on an AID iSPOT plate reader and analysed by automated spot counting.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.