DNA templates
For DNase I footprinting experiments, a 2.8 kb plasmid containing 360 bp of telomeric DNA was amplified in SURE2 Escherichia coli cells grown at 30 °C and extracted using a QIAGEN Plasmid Maxi kit. The plasmid was linearized by BsmFI digestion for 1 h at 37 °C leaving one end with telomeric TTAGGG repeats. The DNA was dephosphorylated with Quick-CIP for 30 min at 37 °C and cleaned up by phenol chloroform extraction and ethanol precipitation. The DNA was subsequently phosphorylated using PNK and [γ-32P]ATP for 1 h at 37°C, passed over a G-50 desalting column and phenol chloroform extracted. Labelled DNA was digested with SacI for 1 h at 37 °C to isolate a 390-bp DNA fragment ending in 60 TTAGGG repeats from the rest of the plasmid. Digested DNA was run on a 10% TBE-PAGE gel (Invitrogen) after which the telomeric fragment was excised and gel extracted by shaking in 10 mM Tris pH 8.0, 300 mM NaCl, 1 mM EDTA overnight at 21 °C. The final DNA fragment (PE1; see Supplementary Table 1 for sequence) was precipitated with isopropanol and resuspended in 1× TE buffer. A non-telomeric DNA fragment (PE2; see Supplementary Table 1 for sequence) containing 360 bp of random DNA sequence in place of TTAGGG repeats was prepared through the same procedure using a non-telomeric plasmid. Telomeric DNA templates for experiments in Extended Data Fig. 2c were prepared as above but with initial digestion by Esp3I instead of BsmFI, thereby yielding a different DNA end compatible with ssDNA overhang ligation. To prepare a 15-nt overhang template, dephosphorylated DNA was additionally mixed with oligonucleotide PE5 at a 1:75 DNA:oligonucleotide molar ratio and incubated overnight at 16 °C with T4 DNA ligase (NEB). The ligated template was subsequently cleaned up from excess oligonucleotide using two rounds of HighPrep PCR cleanup beads (MAGBIO), resuspended in TE buffer and γ-32P-labelled as above.
Telomeric DNA substrates for cryo-EM, crosslinking and DNA-PK pulldown experiments were prepared by mixing oligonucleotides PE3 and PE4 at a 1:1 molar ratio, heating to 95 °C and cooling to room temperature over 2 h.
Protein expression
Open reading frames for human KU70/80, RAP1 or TRF2 were codon-optimized for Spodoptera frugiperda and cloned into pACEBAC1 vector. The mutants indicated were generated using PCR-based mutagenesis (see Supplementary Table 2 for mutant details). The shelterin genes (TERF1, TERF2, RAP1, TINF2, ACD (also known as TPP1) and POT1), codon-optimized for E. coli, were synthesized by GenScript and cloned into pACEBAC1, using a nicking cloning system43. Vectors were transposed into DH10 MultiBac Competent E. coli cells and grown in LB medium overnight at 37 °C shaking at 200 rpm. Bacmid DNA was extracted and used to transfect Sf9 insect cells which were subsequently grown at 27 °C, shaking at 130 rpm over several cell passages. At passage three (P3), 200 ml of High Five insect cells at a 1.5 × 106 density were infected with baculoviruses for protein expression, incubating at 27 °C with 130 rpm. After 3 days, the cell count was checked, and cells were harvested by centrifugation at 760g for 20 min at 4 °C. Cell pellets were then resuspended in PBS, transferred into 50-ml Falcon tubes and pelleted again by centrifugation at 470g for 25 min. Supernatants were discarded, the pellets flash frozen in liquid nitrogen and placed at -80 °C until required.
Nuclear extract preparation
HeLa cell pellets were resuspended in buffer A (10 mM HEPES pH 8.0, 10 mM KCl, 1.5 mM MgCl2, 0.5 mM DTT, 0.5 mM AEBSF) and incubated for 10 min at 4 °C. Following centrifugation at 1,033g for 10 min at 4 °C, the pellet was resuspended in two pellet volumes of buffer A and lysed by 20 strokes in a Dounce homogenizer (pestle type B). Lysate was centrifuged at 25,000g for 20 min at 4 °C and the pellet (containing nuclei) was resuspended in 1.3× pellet volumes of buffer C (20 mM HEPES pH 8.0, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 25% glycerol, 0.5 mM DTT, 0.5 mM AEBSF). After 20 strokes in a Dounce homogenizer (pestle type B), nuclei were incubated for 30 min at 4 °C and centrifuged at 25,000g for 30 min at 4 °C. The supernatant was collected and flash frozen in liquid nitrogen.
Protein purification
DNA-PKcs
HeLa cell nuclear extract was diluted into DPKQ buffer (20 mM HEPES pH 7.6, 100 mM NaCl, 2 mM MgCl2, 0.5 mM EDTA, 10% glycerol, 0.5 mM DTT, 0.5 mM AEBSF) and ultracentrifuged at 50,000g for 1 h at 4 °C. The supernatant was collected and filtered through a 0.45-µm syringe filter before injection onto a Q-sepharose column equilibrated in DPKQ buffer. The column was washed in DPKQ buffer and proteins eluted over a 100 mM–1 M NaCl gradient. Fractions were spotted onto a nitrocellulose membrane and Western blotted to identify DNA-PKcs-containing fractions, which were pooled, diluted into DPKQ buffer and loaded onto a heparin column equilibrated in the same buffer. The column was washed with DPKQ buffer and proteins eluted over a 100 mM–1 M NaCl gradient. DNA-PKcs-containing fractions were pooled, dialysed into DPKC buffer (50 mM Tris pH 7.3, 0.5 mM EDTA, 5% glycerol, 2.5 mM DTT, 0.5 mM AEBSF) with 50 mM KCl, and loaded onto a dsDNA-conjugated CNBr-activated Sepharose column equilibrated in the same buffer. The column was washed in DPKC buffer with 50 mM KCl and bound proteins were eluted in DPKC buffer with 411 mM KCl. DNA-PKcs-containing fractions were pooled, diluted into DPKC buffer with 100 mM KCl and 0.02% Tween-20 and loaded onto a MonoQ column equilibrated in the same buffer. The column was washed in DPKC buffer with 100 mM KCl and 0.02% Tween-20 and proteins were eluted over a 100 mM–1 M KCl gradient. DNA-PKcs fractions were pooled, diluted into DPKC buffer with 100 mM KCl and 0.02% Tween-20 and loaded onto a MonoS column equilibrated in the same buffer. The column was washed in DPKC buffer with 100 mM KCl and 0.02% Tween-20 and proteins were eluted over a 100 mM–1 M KCl gradient. Final DNA-PKcs fractions were pooled, concentrated using a 100 kDa Amicon Ultra centrifugal filter, flash frozen in liquid nitrogen and stored at −80 °C.
KU70/80
Cell pellets were resuspended in 50 mM Tris pH 8.0, 2 mM β-mercaptoethanol with protease inhibitor tablets and incubated stirring for 20 min at 4 °C. Cells were lysed by the addition of 16.7% glycerol and 300 mM NaCl, stirring for 30 min at 4 °C. Lysed cells were ultracentrifuged at 125,000g for 30 min at 4°C and the supernatant was incubated with anti-Flag resin for 2 h at 4 °C. Beads were successively washed in Ku buffer (50 mM Tris pH 8.0, 5% glycerol, 2 mM β-mercaptoethanol) with 300 mM NaCl followed by Ku buffer with 150 mM NaCl. Proteins were eluted using the latter buffer supplemented with 0.5 mg ml−1 3× Flag peptide. Proteins were subsequently separated on a Superdex 200 gel filtration column, equilibrated and run using Ku buffer with 150 mM NaCl. Final Ku70/80 fractions were pooled, concentrated using a 30 kDa Amicon Ultra centrifugal filter, flash frozen in liquid nitrogen and stored at −80 °C.
TRF2 and RAP1
For TRF2 and RAP1, cell pellets were resuspended in 50 mM HEPES pH 7.6, 1 mM DTT with EDTA-free protease inhibitor tablets (one per 50 ml, Roche) and incubated with stirring for 20 min at 4 °C. Protein was extracted by the addition of 16.7% glycerol and 300 mM NaCl, stirring for 30 min at 4°C. Extract was cleared by ultracentrifugation at 125,000g for 30 min at 4 °C. The supernatant was applied to Strep-Tactin XT 4Flow resin equilibrated in shelterin buffer (50 mM HEPES pH 7.6, 500 mM NaCl, 10% glycerol, 1 mM DTT). Beads were washed in shelterin buffer and proteins were eluted with the same buffer supplemented with 10 mM biotin. Proteins were subsequently separated on a Superdex 200 10/300 column equilibrated and run in shelterin buffer. Final TRF2 or RAP1 fractions were pooled, concentrated using a 30 kDa Amicon Ultra centrifugal filter, flash frozen in liquid nitrogen and stored at −80 °C. To cleave the TEB1–RAP1 fusion protein, 0.24 µM PreScission protease was incubated with 1.2 µM TEB1–RAP1 for 2 h at 4 °C prior to experiments.
Shelterin
For the shelterin complex with and without POT1–TPP1, cell pellets containing all 6 shelterin subunits or all subunits except POT1 and TPP1 overexpressed were resuspended in lysis buffer (50 mM Hepes pH 8.0, 300 mM NaCl, 10% glycerol, 1 mM MgCl2, 10 mM beta-mercaptoethanol, 0.1 µl ml−1 Base muncher nuclease, 8 µg ml−1 Avidin, 1 mM AEBSF and EDTA-free protease inhibitor tablets (one per 50 ml, Roche)) and cells were lysed by sonication. Lysate was cleared by centrifugation at 48,380g for 1 h, 4 °C. Cleared lysate was passed through a 0.45-µm filter and applied to a StrepTrap column equilibrated with StrepTrap wash buffer (50 mM Hepes 8.0, 300 mM NaCl, 10 glycerol and 1 mM tris-2-carboxyethyl phosphine (TCEP)), which was washed with 20 column volumes StrepTrap wash buffer prior to elution with 5 column volumes StrepTrap wash buffer supplemented with 10 mM d-desthiobiotin. Eluate (the 0.5 ml eluate containing the highest concentration of shelterin) was applied to a Superose 6 10/300 column preequilibrated in 50 mM Hepes pH 8.0, 300 mM NaCl, 10% glycerol and 1 mM TCEP, and run in the same buffer. The desired fractions were aliquoted and flash frozen for storage. Further characterization of the full shelterin complex will be reported elsewhere.
XRCC4–LIG4
Cell pellets were resuspended in 50 mM HEPES pH 7.6, 1 mM DTT, 1 mM EDTA with EDTA-free protease inhibitor tablets (Roche, 1 per 50 ml buffer) and incubated with stirring for 20 min at 4 °C. Proteins were extracted by the addition of glycerol to 16.7% and NaCl to 300 mM with stirring for 30 min at 4 °C. Extract was centrifuged at 41,656g for 30 min at 4 °C and the supernatant was applied to Strep-Tactin XT 4Flow resin equilibrated in X4 buffer (50 mM HEPES pH 7.6, 300 mM NaCl, 10% glycerol, 1 mM DTT, 1 mM EDTA). Beads were washed in X4 buffer and proteins were eluted with the same buffer supplemented with 30 mM biotin. Protein fractions were pooled, the NaCl concentration diluted to 100 mM then applied to a 1 ml heparin column and subjected to a linear gradient from 0.1 M to 1 M NaCl over 20 column volumes. Proteins were subsequently separated on a Superdex 200 gel filtration column equilibrated and run in 100 mM NaCl buffer (50 mM HEPES pH 7.6, 100 mM NaCl, 10% glycerol, 1 mM DTT, 1 mM EDTA). Final XRCC4–LIG4 fractions were pooled, concentrated using a 30 kDa Amicon Ultra centrifugal filter, flash frozen in liquid nitrogen and stored at −80 °C.
Nano differential scanning fluorimetry
Purified proteins as indicated were analysed by nano differential scanning fluorometry to assess thermal protein stability using a Tycho NT.6 (Nanotemper) with 10 µl capillaries, monitoring fluorescence at 330 and 350 nm over a 35–95 °C temperature ramp (30 °C min−1).
DNase I footprinting
[γ-32P]-labelled PE1 (telomeric) or PE2 (non-telomeric) template (2 nM) was mixed with 30 nM DNA-PKcs and 50 nM KU70/80 in 25 mM HEPES pH 7.6, 80 mM KCl, 1.5 mM CaCl2, 1.5 mM MgCl2, 5% glycerol, 50 µg ml−1 BSA, 2 mM DTT and incubated on ice for 5 min. 5 nM shelterin or TRF2–RAP1 (dimer concentration of TRF2) was added to DNA-bound DNA-PK and incubated at 37 °C for 10 min. Nuclease cleavage was initiated by addition of DNase I to 0.5 U ml−1 and the reactions were incubated for a further 2 min at 37 °C before quenching with 25 mM EDTA, 0.2% SDS, 0.2 mg ml−1 Proteinase K. Following incubation at 37 °C for 10 min, samples were extracted with phenol chloroform, ethanol precipitated and resuspended in 2 µl 99% formamide, 5 mM EDTA, bromophenol blue. Samples were boiled for 2 min and run on an 8% urea-PAGE sequencing gel in 1× TBE. Gels were subsequently dried and exposed to a BAS-MS imaging plate before phosphor imaging using a Typhoon Biomolecular Imager (Amersham). Images were analysed in ImageJ2 (v.2.14.0) and Adobe Photoshop (v.25.0.0) and figures were prepared using Adobe Illustrator (v.25.0.0). Footprinting experiments in Extended Data Fig. 2b,c were prepared as above but with higher protein and/or DNA concentrations as indicated in figure legend. Extended Data Fig. 2c was also performed with coincident addition of shelterin, KU and DNA-PKcs.
Electrophoretic mobility shift assay
[γ-32P]-labelled PE1 (telomeric) template (1 nM) was mixed with 2, 15 or 40 nM TRF2 in 25 mM HEPES pH 7.6, 80 mM KCl, 1.5 mM CaCl2, 1.5 mM MgCl2, 5% glycerol, 50 µg ml−1 BSA, 2 mM DTT. 10 µl reactions were incubated at 37 °C for 15 min. Samples were supplemented with 1% sucrose, Orange G and run on a 1.5% agarose gel in 0.5× TBE. Gels were dried and analysed by phosphor imaging as with DNase I footprinting experiments.
Crosslinking
KU70/80 (200 nM) was mixed with 200 nM RAP1 in 20 mM HEPES pH 7.6, 200 nM NaCl, 2 mM MgCl2, 0.5 mM EDTA, 10% glycerol, 0.5 mM DTT, 0.5 mM AEBSF and incubated for 5 min at 4 °C. Samples were supplemented by 100 nM annealed PE3/PE4 DNA substrate and incubated for 10 min at 4 °C. Proteins were crosslinked by addition of 2 mM DSSO and incubated for 60 min at room temperature. Reactions were stopped with 20 mM Tris pH 7.6. Samples were run on a Criterion XT 3-8% Tris-Acetate PAGE gel in 1× XT Tricine and analysed by silver staining (SilverQuest, Invitrogen) or immunoblotting, probing for KU70 (Flag) or RAP1 (strep) (see ‘Antibodies’ and ‘Immunoblotting’). The crosslinking experiment in Extended Data Fig. 2i was performed as above with 200 nM TRF2(ΔBΔTRFH) (with cleaved off strep tag) added together with RAP1.
DNA-PK pulldown
Annealed PE3/PE4 DNA substrate (5 nM) preincubated with a twofold excess of streptavidin (IBA) was incubated with 10 nM KU70/80 and 15 nM DNA-PKcs for 3 min at 30 °C in 20 mM HEPES pH 7.6, 80 mM KCl, 5% glycerol, 0.01% NP-40 and 1 mM DTT in a final volume of 25 µl in protein low bind tubes (Alpha Labs). 5 nM TRF2–RAP1 complex (dimer concentration of TRF2) was added, and after a further 3 min, 30 nM XRCC4–LIG4 complex was added. After a further 3 min, the complete reaction was added to 1 µl equivalent of anti-Flag M2 magnetic beads (Sigma), and the mixture incubated at 4 °C with shaking for 30 min. Beads were pelleted on a magnetic rack, washed 3× with 50 µl 25 mM HEPES pH 7.6, 80 mM KCl, 10% glycerol, 0.02% NP-40 and 1 mM DTT with a brief vortex included for each wash. Beads were resuspended in wash buffer supplemented with 0.25 mg ml−1 3× Flag peptide and incubated for 20 min at 18 °C with shaking. Eluted proteins were supplemented with SDS loading buffer, run on a 4–12% TGX precast gel (Bio-Rad), transferred onto nitrocellulose membrane at 80 V for 90 min and detected by immunoblotting with the antibodies indicated (see ‘Antibodies’ and ‘Immunoblotting’). Pulldown experiments in Extended Data Fig. 7b,c were prepared as above but with 30–120 nM XRCC4–LIG4 as indicated.
Cryo-EM sample preparation
Annealed PE3/PE4 DNA substrate (see ‘DNA templates’) was mixed 1:1 with streptavidin in TE buffer and incubated for 30 min at room temperature. Streptavidin-bound DNA was diluted to 250 nM in 40 mM HEPES pH 7.6, 100 mM NaCl, 3 mM MgCl2, 1 mM DTT, 2.5% glycerol and incubated with 250 nM KU70/80 and 250 nM DNA-PKcs for 10 min at 4 °C. 250 nM TRF2–RAP1 (dimer concentration of TRF2) was added and incubated for another 5 min at 4 °C. Samples were supplemented with 0.05% CHAPS prior to cryo-EM grid preparation.
Cryo-EM data acquisition and image processing
Copper R1.2/1.3 grids (300-mesh, Quantifoil) were coated with a thin layer or carbon and glow-discharged at 15 mA for 30 s (PELCO easiGlow). Three microlitres of sample was applied to glow-discharged grids and incubated for 5 s followed by blotting for 3 s using a Vitrobot Mark IV (Thermo Scientific) operated at 4 °C and 100% humidity. Grids were subsequently plunge-frozen in liquid ethane. Cryo-EM data were acquired at 200 kV on a Glacios Cryo-TEM (Thermo Scientific) equipped with a Falcon 4i Direct Electron Detector (Thermo Scientific). In total, 30,604 movies with 30 frames were collected at 150,000× magnification (0.94 Å pixel size) with a total electron dose of 50 e− Å−2 and a defocus range of −1.0 to −2.5 μm (see Extended Data Table 1). Subsequent image processing was performed in cryoSPARC (v4.3.1)44. Movies were motion corrected using patch alignment with all frames followed by patch contrast transfer function estimation. Particles were picked through automated template-based picking (template EMD-6803) and extracted with 4× binning using a box size of 96 pixels. Following 2D classification 1,464,362 DNA-PK particles were selected to reconstruct an ab initio 3D model, subsequently used as a starting model for heterogeneous refinement using 5 classes. The most prominent DNA-PK class was selected and subjected to homogeneous refinement followed by local refinement using a focus mask encompassing the KU70/80–DNA core. A 3D classification without alignment using the same mask and ten classes was performed to identify particles containing additional RAP1 and KU densities. Classes lacking either the RAP1 BRCT the RAP1 Myb domain or the KU70 SAP domain were excluded. A total of 526,885 selected particles was re-extracted unbinned with a 384-pixel box size and a 3D map was reconstructed through homogeneous refinement. Following global contrast transfer function refinement a structure of the full DNA end-binding complex was resolved to 3.58 Å using homogeneous refinement. The KU–RAP1–DNA core was locally refined to 3.32 Å using a focus mask excluding DNA-PKcs. The DNA-PKcs–DNA density was similarly refined using a mask excluding the KU–RAP1–DNA core and subjected to 3D classification without alignment using the same mask. Some flexibility in DNA-PKcs conformation was observed and classes of the most prominent conformation containing 370,172 particles were selected. A DNA-PKcs–DNA structure from these particles was resolved to 3.40 Å by homogeneous refinement followed by local refinement using a DNA-PKcs focus mask. Maps for the full end-binding complex and the locally refined densities were sharpened in cryoSPARC and combined using Phenix (v1.20.1) combine_focused_maps45. The composite map (EMD-19065) was subsequently used for model building in Coot46 and figure generation in ChimeraX47.
Model building and validation
Molecular models for human DNA-PK (PBD 7K1K), RAP1 Myb (PDB 1FEX) and KU70 SAP (PDB 1JJR) were docked into the cryo-EM map using the Fit in Map command in ChimeraX47. The RAP1 BRCT domain from a KU–RAP1 AlphaFold model (see ‘AlphaFold modelling’) was similarly docked into the cryo-EM map. Models were refined against the map using Namdinator48 followed by manual inspection in Coot46. Unoccupied protein densities and nucleic acids were modelled de novo. The resulting model was iteratively refined using Phenix (v1.20.1) real_space_refinement49 with geometry and secondary structure restraints followed by manual adjustment in Coot. The quality of the final atomic model (PDB 8RD4) was evaluated by MolProbity50 in Phenix (see Extended Data Table 1).
AlphaFold modelling
Full-length human RAP1, KU70 and KU80 were analysed using AlphaFold 3 on the online AlphaFold server, with the top ranked prediction shown. For Extended Data Fig. 9, the additional sequences analysed were as follows. Salmo salar: NP_001133439.1, XP_014030197.1 and XP_045561280.1; Strongylocentrotus purpuratus: XP_030845408.1, XP_030843748.1, XP_001198957.2; Nematostella vectensis: XP_001641354.1, EDO36674.1, EDO44451.1; Trichoplax adhaerens: XP_002117640.1, XP_002117043.1, XP_002112721.1.
Protein alignments
Pre-computed protein alignments were analysed using the ProViz online tool51.
Antibodies
Human DNA-PKcs was detected with antibody 18-2 (Invitrogen MA5-13238) at 1:100 dilution, human RAP1 with antibody A300-306A (Bethyl Laboratories) at 1:4,000 dilution, human TRF2 with antibody D1Y5D (Cell Signaling 13136) at 1:1,000 dilution, human Apollo with antibody HPA064934 (Atlas Antibodies) at 1:100 dilution and human α-tubulin with antibody T9026 (Sigma) at 1:1,000 dilution. Recombinant human KU70 was detected via an N-terminal Flag tag with antibody M2 (Sigma F1804) at 1:1,000 dilution. Recombinant human LIG4, TRF2 and RAP1 were detected via a dual strep tag using antibody ab76949 (Abcam) at 1:1,000 dilution. Recombinant human XRCC4 was detected with antibody C-4 (Santa Cruz sc-271087) at 1:500 dilution. Primary antibodies for human proteins were detected with goat anti-rabbit IgG–horseradish peroxidase (HRP) (Cell Signalling 7074) or horse anti-mouse IgG–HRP (Cell Signalling 7076) secondary antibody. Mouse RAP1 was detected with antibody D9H4 (Cell Signalling 5433) at 1:1,000 dilution, mouse TRF2 with antibody D1Y5D (Cell Signaling 13136) at 1:1,000 dilution and mouse β-actin with antibody 8H10D10 (Cell Signaling 3700) at 1:4,000 dilution followed by donkey anti-rabbit IgG–HRP (NA934V, Cytiva), goat anti-rabbit IgG–HRP (31460, Invitrogen) or goat anti-mouse IgG–HRP peroxidase (31430, Invitroen) secondary antibody.
Cell lines and viral gene delivery
SV40-LT Apollofl/fl Lig4+/+, Apollofl/fl Lig4−/− and Trf2fl/flRosa26cre-ERT1 MEFs have been previously described10,52. hTERT immortalized RPE-1 cells have been previously described53. All MEFs were immortalized with pBabeSV40LargeT and cultured in Dulbecco’s Modified Eagle Medium (DMEM, Cytiva) supplemented with 15% fetal bovine serum (FBS, Gibco), non-essential amino acids (Cytiva), l-glutamine (Cytiva), penicillin-streptomycin (Cytiva), 50 µM β-mercaptoethanol (Sigma). 293 T and Phoenix eco cells (ATCC) were cultured in DMEM (Cytiva) supplemented with 10% HyClone Bovine Calf Serum (Cytiva), non-essential amino acids (Cytiva), l-glutamine (Cytiva), and penicillin-streptomycin (Cytiva). RPE-1 cells were cultured in DMEM/F12 medium supplemented with 10% (v/v) FBS, 1% (v/v) penicillin-streptomycin, 1% Glutamax, 0.5 µg ml−1 Amphotericin B and 0.26% sodium bicarbonate. To generate TP53−/− RPE-1 clones by CRISPR–Cas9 mediated gene editing, cells were electroporated with Cas9–sgRNA ribonucleoparticles targeting the sequences 5′-AAATTTGCGTGTGGAGTATT-3′ and 5′-TCCACTCGGATAAGATGCTG-3′54 using the Neon Transfection system as described55. After 4 days, single cells were sorted into 96-well plates containing medium supplemented with 10 µM nutlin-3a. After 14 days, surviving clones were expanded, and p53 status was analysed by immunoblotting and sequencing of the TP53 locus as described54. CRISPR–Cas9 mediated editing of human RAP1 in TP53−/− RPE-1 cells was carried out using phosphorothioated single-stranded DNA repair templates (ssODN) and selection for positive integrands by ouabain as described56,57. In brief, RAP1 guide RNA 5′-GGCCCAGCCCGGCCAAGCGT-3′ was cloned into the BspI site of Addgene vector 86613. Repair templates for ATPA1 and RAP1 were synthesized by Integrated DNA Technologies (IDT) with the following sequences: ATP1A1: C*A*ATGTTACTGTGGATTGGAGCGATTCTTTGTTTCTTGGCTTATAGCATCAGAGCTGCTACAGAAGAGGAACCTCAAAACGATGACGTGAGTTCTGTAATTCAGCATATCGATTTGTAGTACACATCAGATATC*T*T; RAP1: C*A*TTCCTCGACTCTGTTCGTGAGGGACGACGGCAGCTCCATGTCCTTCTACGTGCGGCCCAGCCCGGCCGACGAGCGCCTCTCGACGCTCATCCTGCACGGCGGCGGCACGGTGTGCGAGGTGCAGGAGCCCGGGGCCGTGCTGCTGGCCCAGCCCGGGGAGGCGCTGGCCGAGGCCTCGGGTGATTTCATCTCCACG*C*A, where * denotes a phosphorthiolated base. To generate RAP1(KR/DE) clones, 300,000 TP53−/− RPE-1 cells were electroporated with the Neon Transfection System using a 10 µl tip and two pulses at 1,350 V and 20 ms with 500 ng RAP1 guide RNA/Addgene plasmid #86613, 2 pmol of ATP1A1 ssODN and 6 pmol RAP1 ssODN. To generate RAP1−/− clones, the procedure was repeated omitting RAP1 ssODN. After 3–4 days, cells were expanded into 15-cm dishes and treated with 0.25 µM ouabain, followed by isolation of single clones 7–12 days after drug selection. Genomic DNA was prepared using EZNA Tissue DNA kit according to the manufacturer’s instructions. RAP1 was amplified by PCR using oligonucleotide sequences AGTGCTGCGCTTCGCGGC and CGCCTTCCGCTTGAGCTTCTG. Editing was analysed by restriction digestion with SalI and Sanger sequencing, and positive clones were single cell sorted and expanded prior to freezing. For retro or lentiviral transduction, a total of 20 µg of plasmid DNA was transfected into Phoenix eco or 293 T cells, respectively, using CaPO4 precipitation. The viral supernatant was filtered through a 0.45-μm filter, supplemented with 4 μg ml−1 polybrene, and used for the transduction of target cells. Lentiviral particles containing the sgRNA against mouse Rap1 (target: 5′-GCAGTCTAGGATGTACTGCG-3′) in lentiCRISPR v2 (Addgene plasmid #52961, a gift from F. Zhang) were introduced into target MEF cells with three infections per day (6–12 h intervals) over 2 days, followed by 2 days in 2–4 µM Puromycin or 340 µM Hygromycin. The same approach was used to target human Apollo in TP53−/− RPE-1 cells with lentiviral particles containing lentiCRISPR v2 with the guide sequence 5′-CTGGTTCCAACGCAGCATGT-3′23, or non-targeting control sequence 5′-CGCCAAACGTGCCCTGACGG-3′. For MEF experiments, Cre was induced with three infections per day (6–12 h intervals) over two days with pMMP Hit&Run Cre retrovirus produced in Phoenix eco cells. Time-point 0 was set 12 h after the first Hit&Run Cre infection. For the Rap1-complementation assay, Apollofl/fl MEFs were transduced with retroviral particle containing Rap1, Rap1KR/DE or Rap1R130E cloned in Plpc vector for a total of 4 infections at 6–12-h intervals, selected for 2–3 days in 2–4 µM Puromycin, transduced with the sgRNA against mouse Rap1 cloned in LcV2-Hygro (Addgene plasmid #91977, a gift from J. Mendell), selected for 2 days in 340 µM Hygromycin and then transduced with pMMP Hit&Run Cre as previously described. For the Trf2-complementation assays, Trf2fl/flRosa26cre-ERT1 MEFs were transduced with retroviral particle containing the Trf2 mutants, selected for 2 days in 2–4 µM Puromycin and then treated with 1 μM 4-OHT for 24 h. Time-point 0 was set at the time of 4-OHT addition. All cell lines in this study routinely tested negative for mycoplasma contamination. RPE-1 cells were validated by whole-genome sequencing, while MEFs were automatically genotyped after isolation by TransnetYX for the presence of Apollo or Trf2 flox alleles and/or ligase 4 deletion or RsCre.
Generation of Trf2-mutant alleles
PCR was used to delete the RAP1-binding motif (RBM) or insert the S367A mutation into MYC-tagged Trf2, Trf2F120A, Trf2ΔiDDR and Trf2F120A ΔiDDR alleles cloned in pLPC retroviral vector13 using the following primers: Trf2ΔRBM-F: 5′-AATCTGGCATCCCCATCATCAC-3′; Trf2ΔRBM-R: 5′-TCTGCTTGGAGGCTCTCTAAG-3′; Trf2S367A-F: 5′-GCGCCAGCCCACAAACACAAGAGACC-3′; Trf2S367A-R: 5′-TGATGGGGATGCCAGATTAGCAAG-3′.
Fluorescence in situ hybridization
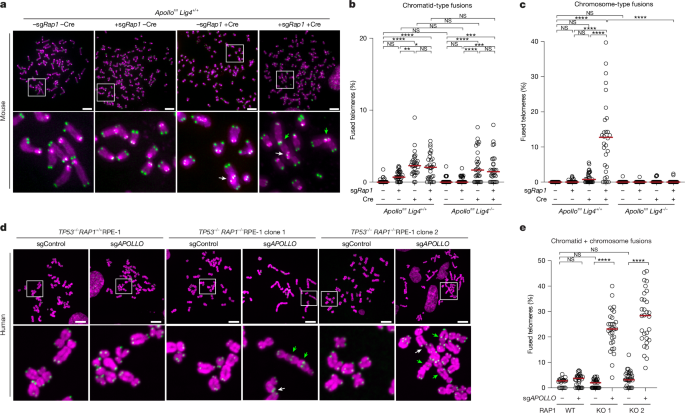
Telomere FISH on mouse and human cells was performed as previously described58. In brief, cells were treated with 0.2 µg ml−1 Colcemid (Biowest/Roche) for 2 h before collection by trypsinization. Collected cells were swollen in a hypotonic solution of 75 mM KCl at 37 °C for 10–20 min before fixation in methanol:acetic acid (3:1) overnight at 4 °C. Cells were dropped onto glass slides and allowed to age overnight. The slides were then dehydrated through an ethanol series of 70%, 95% and 100% and allowed to air dry. Telomere ends were hybridized with Cy3-OO-(TTAGGG)3 in hybridization solution (70% formamide, 1 mg ml−1 blocking reagent (1109617601, Roche), and 10 mM Tris-HCl pH 7.2) for 2 h following an initial 5–10 min denaturation step at 80 °C, washed twice with 70% formamide; 0.1% BSA; 10 mM Tris-HCl, pH 7.2 for 15 min each, and thrice in 0.08% Tween-20; 0.15 M NaCl; 0.1 M Tris-HCl, pH 7.2 or PBS for 5 min each. For MEFs, chromosomal DNA was counterstained with the addition of DAPI (D1306, Invitrogen) to the second wash. Slides were left to air dry and mounted in antifade reagent (Prolong Gold Antifade P36934, Fisher). For RPE-1 cells, air-dried slides were mounted in DAPI-supplemented Vectashield mounting medium (Vector laboratories) Micrographs for mouse cell experiments were collected on a DeltaVision RT microscope, Micrographs for RPE-1 cell experiments were collected on a Zeiss Axio Observer Z1 Marianas TM microscope (operated with 3i SlideBook) equipped with a CSU-X1 spinning disk (Yokogawa). Metaphases were analysed in FIJI and scored fusions were plotted using GraphPad Prism. Figures were prepared in Adobe Illustrator (v25.0.0). CO-FISH analysis of RPE-1 cells was performed as previously described23 with the following modifications: RPE-1 cells were treated with BrdU:BrdC for 14 h, slides were treated with UV for 10 min using a Blak ray model UV-21 365 nm handheld lamp at a distance of 8 cm and telomeres were detected with Cy3-OO-(TTAGGG)3 and FITC-OO-(CCCTAA)3.
Immunoblotting
Cells were lysed in 2× Laemmli buffer at 5 × 103 or 1 × 104 cells per μl and the lysate was denatured for 10 min at 95 °C before shearing with an insulin needle or sonication. Lysate equivalent to 1–2 × 105 cells was resolved using SDS–PAGE and transferred to a nitrocellulose membrane. For mouse cell experiments, Western blot was performed with 5% milk in PBS containing 0.1% (v/v) Tween-20 (PBS-T). For RPE-1 cell experiments, Western blotting was performed in TBS buffer supplemented with 0.1% Tween-20. Additional reagents are described in the ‘antibodies’ section. Immunoblots were developed using chemiluminescence western blotting detection reagents (Cytiva or Cell Signalling or Millipore) and imaged on a ChemiDoc (Bio-Rad) imaging system or using Amersham Hyperfilm MP (Cytiva) and a CURIX 60 processor (AGFA). Images were analysed in Adobe Photoshop (v25.0.0) and figures were prepared using Adobe Illustrator (v25.0.0).
Chemical crosslinking mass spectrometry analysis
Complex assembly and crosslinking with DSSO was performed as described in ‘Crosslinking’. After the crosslinking reaction, triethylammonium bicarbonate buffer (TEAB) was added to the sample at a final concentration of 100 mM. Proteins were reduced and alkylated with 5 mM TCEP and 10 mM iodoacetamide simultaneously and digested overnight with trypsin at final concentration 50 ng μl−1 (Pierce). Sample was dried and peptides were fractionated with high-pH Reversed-Phase (RP) chromatography using the XBridge C18 column (1.0 × 100 mm, 3.5 μm, Waters) on an UltiMate 3000 HPLC system. Mobile phase A was 0.1% v/v ammonium hydroxide and mobile phase B was acetonitrile, 0.1% v/v ammonium hydroxide. The peptides were fractionated at 70 μl min−1 with the following gradient: 5 min at 5% B, up to 15% B in 3 min, for 32 min gradient to 40% B, gradient to 90% B in 5 min, isocratic for 5 min and re-equilibration to 5% B. Fractions were collected every 100 s, SpeedVac dried and pooled into 12 samples for mass spectrometry analysis. Liquid chromatography–mass spectrometry analysis was performed on an UltiMate 3000 UHPLC system coupled with the Orbitrap Ascend Mass Spectrometer (Thermo). Each peptide fraction was reconstituted in 30 μl 0.1% formic acid and 15 μl were loaded to the Acclaim PepMap 100, 100 μm × 2 cm C18, 5 μm trapping column at 10 μl min−1 flow rate of 0.1% formic acid loading buffer. Peptides were then subjected to a gradient elution on the Acclaim PepMap (75 μm × 50 cm, 2 μm, 100 Å) C18 capillary column connected to the EASY-Spray source at 45 °C with an EASY-Spray emitter (Thermo, ES991). Mobile phase A was 0.1% formic acid and mobile phase B was 80% acetonitrile, 0.1% formic acid. The gradient separation method at flow rate 300 nl min−1 was as follows: for 80 min gradient from 5–35% B, for 5 min up to 95% B, for 5 min isocratic at 95% B, re-equilibration to 5% B in 5 min, for 5 min isocratic at 5% B. Precursors between 380 and 1,400 m/z and charge states 3–8 were selected at 120,000 resolution in the top speed mode in 3 s and were isolated for stepped HCD fragmentation (collision energies (%) = 21, 27, 34) with quadrupole isolation width 1.6 Th, Orbitrap detection with 30,000 resolution and 70 ms maximum injection time. Targeted mass spectrometry precursors were dynamically excluded from further isolation and activation for 45 s with 10 ppm mass tolerance. Identification of crosslinked peptides was performed in Proteome Discoverer 2.4 (Thermo) with the Xlinkx search engine in the MS2 mode for DSSO/+158.004 Da (K). Precursor and fragment mass tolerances were 10 ppm and 0.02 Da respectively with maximum 2 trypsin missed cleavages allowed. Carbamidomethyl at C was selected as static modification. Spectra were searched against a UniProt FASTA file containing Homo sapiens reviewed entries. Crosslinked peptides were filtered at FDR < 0.01 using the percolator node and target–decoy database search.
Statistics and reproducibility
Three biological replicates were performed for metaphase spreads, except for those in Extended Data Fig. 1h,i, which were repeated twice. All other experiments were independently replicated a minimum of three times except for Fig. 3c and Extended Data Figs. 2f,g and 7c which were repeated twice. All attempts at replication were successful.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.