Cell lines
All cell lines are listed in Supplementary Table 2 and were cultured at 37 °C with 5% CO2 in DMEM GlutaMAX (Gibco), supplemented with 10% fetal calf serum (Avantor, VWR; Supplementary Table 3) and penicillin–streptomycin (Sigma; Supplementary Table 3).
Compounds and materials
All compounds and instruments are listed in Supplementary Table 3.
Generation of knockout cells
Cells were transfected with Cas9-2A-GFP (Addgene, 48138) containing a guide RNA targeting the gene of interest (sgRNAs are listed in Supplementary Table 4 and plasmids in Supplementary Table 5) using Lipofectamine 2000 (Life Technologies, 11668027). Cells were sorted by fluorescence-activated cell sorting on BFP and GFP and were plated at a low density, after which individual clones were isolated. Isolated knockout clones were verified by Sanger sequencing and/or western blot analysis (primers and antibodies are listed in Supplementary Tables 6 and 7, respectively).
PGK plasmids with GFP-tagged protein
The CFAP20 gene was amplified from cDNA by PCR and inserted in PGK-EGFP-C1-IRES-PURO, thereby tagging CFAP20 at its N terminus with GFP (primers and plasmids are listed in Supplementary Tables 5 and 6). The CFAP20R100C mutant was generated using site-directed mutagenesis PCR. The CCNC gene was amplified from the CMV-EGFP-CCNC plasmid and inserted in pLenti-PGK-GFP-puro, thereby tagging CCNC at its N terminus with GFP. The CCNCD182A mutant was generated using site-directed mutagenesis PCR. A region spanning the CMV promoter was amplified by PCR and used to replace the PGK promoter in pLenti-PGK-GFP-puro. A fragment encoding RNaseH1 from plasmid pFRT-TO-EGFP-RNaseH1 was amplified by PCR and inserted in pLenti-CMV-GFP-puro. All sequences and plasmids were verified by Sanger sequencing.
Generation of stable cell lines
Cells were transfected with Lipofectamine 2000 (Life Technologies, 11668027) or polyethyleneimine reagent (Brunschwig Chemie, 23966-2) according to the manufacturer’s instructions. All plasmids are listed in Supplementary Table 5. Lentiviral particles were produced by co-transfecting pLenti plasmids with pMDLg-pRRE, pRSV-REV and pCMV-VSVG in a 2:1:1:4 ratio in HEK293T cells by using polyethylenimine reagent. After production, lentivirus was filtered through a 0.44-µm filter and added to RPE1 cells in a complete DMEM medium supplemented with 4 µg ml−1 polybrene and 10 mM HEPES. After overnight incubation, the medium was removed, and fresh medium was added. The expression of GFP was verified three days after lentiviral transduction.
CRISPR–Cas9 gene editing
For CRISPR–Cas9 gene editing, we used a previously described approach50. In brief, Cas9 expression was induced by 200 ng ml−1 DOX followed by transfection with 20 nM equimolar crRNA:tracrRNA duplexes with 1:1,000 RNAiMAX (Life Technologies).
RNA interference
For RNA interference (Supplementary Table 4), cells were transfected with 50 nM siRNA duplexes using Lipofectamine RNAiMAX (Invitrogen). Cells were transfected twice with siRNAs at 0 h and 24 h and were typically analysed 60 h after the first transfection.
Zebrafish lines and husbandry
All adult zebrafish strains are listed in Supplementary Table 8 and were raised at 28.5 °C under a 14-h–10-h light–dark cycle. Larvae were raised to 48 hours post-fertilization (hpf) at 28.5 °C in an incubator in E3 medium. The mutant cfap20 line (ua5025)24 was a gift from W. Ted Allison. All fish were on an AB background and staged as previously described51. Anaesthesia for live imaging was achieved with 60 mg l−1 of eugenol. All rescue experiments were performed on at least two clutches. All animal experiments were performed with the approval of the University of Toronto Animal Care Committee in accordance with the guidelines from the Canadian Council for Animal Care (CCAC).
mRNA and morpholino microinjection in zebrafish
All oligonucleotides used in zebrafish strains are listed in Supplementary Table 8. Microinjection into the cell (mRNA) or the yolk syncytial layer (morpholino) before the four-cell stage was done using pulled (P-97; Sutter Instrument) glass capillary tubes (TW100F-4; World Precision Instruments). Unfertilized eggs or embryos stalled during gastrulation were removed at 12 hpf. WT and CFAP20R100C variant mRNAs were transcribed from linearized pCS2+ vectors. The WT-containing plasmid was a gift from W. T. Allison. The R100C variant sequence was ordered as a gBlock (Integrated DNA Technologies) and directionally cloned into pCS2+ using BamHI or XbaI restriction enzymes. In vitro transcription was performed using the SP6 mMessage mMachine kit (Thermo Fisher Scientific, AM1340), followed by phenol:chloroform purification. A dose response using the WT mRNA diluted with ddH2O (into cfap20+/− incross clutches) was performed (25, 100 pg) to ensure that a rescue efficiency higher than 90% was achieved (data not shown). Embryos from cfap20+/− incrosses were microinjected with WT or R100C CFAP20 mRNA and larvae were then raised to 48 hpf and groups were blinded. Larvae were scored on the basis of a straight extension of the anterior–posterior axis (normal) or ventral curling of the body (curvature). Embryos were then processed for DNA lysis and genotyped as below, and groups were then unblinded. Only scores from cfap20−/− homozygotes were analysed. Standard control and ccnc splice morpholino oligonucleotides (Gene Tools), as in a previous study51, were used to knock down ccnc. A dose response of 1, 2 and 4 ng MO was performed in AB incross (2 clutches; more than 20 animals) as in Extended Data Fig. 5f and larvae were scored at 48 hpf on the basis of the severity of the phenotype. An optimal dose of 1.5 ng was chosen for subsequent experiments. Cfap20+/− heterozygote incross embryos were injected, groups were blinded and larvae were raised to 48 hpf. Larvae were scored (as above) on the basis of anterior–posterior curvature (2 clutches; more than 50 animals), then processed for genotyping before unblinding. Only scores from injected or uninjected cfap20−/− homozygotes were analysed.
Zebrafish cfap20 genotyping
A genomic DNA template for PCR was generated by adding tissue to 50 µl 50 mM NaOH, heating at 95 °C for 20 min and then neutralizing with 5.5 µl 1 M Tris-HCl. The template was diluted 50-fold and PCR genotyping was performed using GoTaq 2 (Promega). Primer sequences are listed in Supplementary Table 8.
Microscopic analysis of zebrafish larvae
Larval zebrafish were anaesthetized as above and transferred to 1% agar-lined Petri dishes for imaging. Representative bright-field images were taken using ZEN 3.7 (Zeiss) at 32× magnification on a Lumar V12 (Zeiss) stereomicroscope with an Axiocam 712 mono (Zeiss) camera. All graphing of and statistical tests on zebrafish data were done in Prism 10 (GraphPad), as described in Supplementary Table 9. The absolute number of normal versus axis-curvature defects was compared statistically using Fisher’s exact test. Raw images were cropped, and brightness and contrast were adjusted in Photoshop 2024 (Adobe). Identical transformations were performed on control and experimental images.
Western blotting
Proteins were separated on 4–12% Criterion XT Bis-Tris gels (Bio-Rad, 3450124) in NuPAGE MOPS running buffer (Thermo Fisher Scientific, NP0001–02), or on 3–8% Criterion XT Tris-Acetate protein gel (Bio-Rad, 3450131) in Tris/Tricine/SDS Running Buffer (Bio-Rad, 1610744), followed by blotting onto PVDF membranes (EMD Millipore, IPFL00010). Membranes were blocked with 5% milk powder in phosphate-buffered saline (PBS) with 0.1% Tween for one hour at room temperature. Protein expression was analysed by immunoblotting with the designated primary antibodies (listed in Supplementary Table 7) and corresponding secondary antibodies at 1:10,000. For detection, the Odyssey infrared imaging scanning system (LICORbio) was used.
Immunoprecipitation
Cell pellets were solubilized in EBC-1 (50 mM Tris, pH 7.5, 150 mM NaCl, 0.5% NP-40 and 2 mM MgCl2 with protease inhibitor cocktails (Roche)) supplemented with 500 U benzonase for one hour at 4 °C under rotation. The lysates were cleared from insoluble chromatin by centrifugation and were subjected to immunoprecipitation with GFP Trap beads (Chromotek, GTA-200) for 1.5 h at 4 °C under rotation. The beads were then washed four to six times with EBC-2 buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 0.5% NP-40 and 1 mM EDTA) and boiled in Laemmli buffer. Bound proteins were resolved by SDS–PAGE and immunoblotted with the indicated antibodies (Supplementary Table 7). For endogenous immunoprecipitation, 2 µg of antibody was incubated with the samples in EBC-1 buffer and benzonase, and they were subjected to immunoprecipitation with protein A agarose beads (Millipore, 16-157).
Mass-spectrometry sample preparation
After pull-down, the GFP beads were washed three times with 50 mM ammonium bicarbonate, followed by overnight digestion using 2.5 μg trypsin at 37 °C under constant shaking. Digested peptides were separated from the beads by a 0.45-µm filter column (Meck, UFC30HV00) that was prewashed with 50 mM ammonium bicarbonate. Trypsin activity was quenched by acidifying the sample with trifluoroacetic acid to a final concentration of 1%. Peptides were desalted and concentrated using in-house assembled triple-disc C18 stage-tip columns (serial number 66883-U; Sigma-Aldrich) as previously described52.
Mass-spectrometry data acquisition
The GFP–CCNC and GFP–CCNC(D182A) samples with their corresponding GFP-NLS controls were analysed by on-line C18 nano-high performance liquid chromatography (HPLC) MS/MS with a system consisting of an UltiMate3000 nano gradient HPLC system (Thermo Fisher Scientific) and an Exploris480 mass spectrometer (Thermo Fisher Scientific). Digested peptides were injected onto a cartridge precolumn (300 μm × 5 mm, C18 PepMap, 5 μm) in 100% solvent A (0.1 % formic acid in milli-Q), with a flow of 10 μl per min for 3 min (Thermo Fisher Scientific), and eluted using a homemade analytical nano-HPLC column (30 cm × 75 μm; Reprosil-Pur C18-AQ 1.9 μm, 120 A (Dr. Maisch). The chromatography gradient length was 60 min from 2% to 40% solvent B, followed by a 5-min increase to 95% solvent B, another 5 min of 95% solvent B and back to 2% solvent B for chromatography column reconditioning. The mass spectrometer was operated in positive polarity data-dependent MS/MS mode with a cycle time between master scans of 3 s. Full-scan MS spectra were obtained with a resolution of 60,000, a normalized automatic gain control (AGC) target of 300% and a scan range of 350–1,600 m/z. Precursors were fragmented by higher-energy collisional dissociation (HCD) with a normalized collision energy of 28%. Tandem mass spectra (MS/MS) were recorded with a resolution of 30,000 and a normalized AGC target value of 75%. Precursor ions selected for MS/MS analysis were subsequently dynamically excluded from MS/MS analysis for 30 s and only precursors with a charge state of 2–6 triggered MS/MS events.
Mass-spectrometry data analysis
RAW data were analysed using MaxQuant (v.1.6.14.0) as previously described53,54.
Mass-spectrometry data availability
The mass-spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE55 partner repository with the dataset identifier PXD051449 (GFP–CFAP20(R100C) and GFP–CCNC sample sets).
CRISPR screens
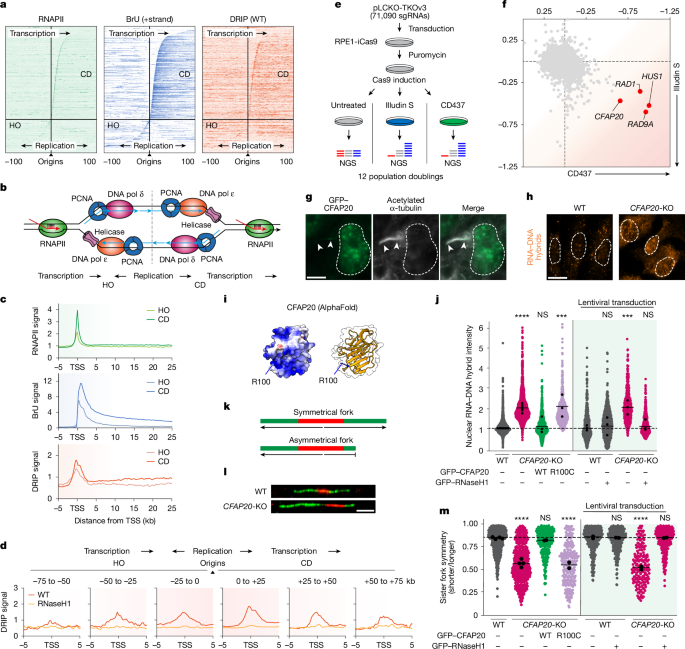
For every screen, three populations of RPE1-iCas9 were transduced at a multiplicity of infection (MOI) of around 0.2 with a 1:1,000 dilution of TKOv3-in-pLCKO lentiviral library in medium containing 8 µg ml−1 hexadimethrine bromide (Sigma-Aldrich). The library was a gift from K. Chan, A. Tong and J. Moffat. Twenty-four hours after transduction, puromycin (Sigma-Aldrich) was added to 5 μg ml−1 to select for transduced cells. After all cells in non-transduced control populations had died and dishes with transduced populations had reached 90% confluence, a t = 0 sample was taken for each of the three populations. From the remaining cells of each population, 30 × 106 (corresponding to a library representation of more than 400) were grown as a control population. To screen for replication stress genes, RPE1-iCas9 parental cells were grown in the presence of the DNA pol α inhibitor CD437 at a concentration of 200 nM. The illudin S screen has been described previously17. To screen for synthetic-viable genes, RPE1-iCas9 CFAP20-KO cells were grown without drugs or inhibitors. DOX was added to the medium of all replicates from t = 0 onwards to induce expression of Cas9, at a concentration of 200 ng ml−1. After 3 doublings, 30 × 106 cells of each population were passed. After 12 doublings, all populations were collected.
Sequencing and analysis of CRISPR screens
Genomic DNA was isolated from each population using the Blood and Cell Culture DNA Maxi Kit (QIAGEN). Then, 3 µg of gDNA from each population was amplified using the KAPA HiFi ReadyMix PCR Kit (Roche) with the TKO outer Fw and Rv primers (primers are listed in Supplementary Table 5), followed by a second PCR reaction using reverse primers with different Illumina i7 index sequences for each sample to identify the sample after pooled sequencing as described56. The second PCR products of each pool were purified using the QIAquick PCR Purification Kit (QIAGEN). Samples were sequenced on a NovaSeq 6000 and reads were mapped to the TKOv3 library sequences, not allowing any mismatches. To compare the illudin S to the CD437 screen (Fig. 1f), the lowest z-score for each screen was normalized to −1 (sensitizer genes:UVSSA for illudin S; HUS1 for CD437), and the highest score was normalized to +1 (resistance genes: PTGR1 for illudin S; CDAN1 for CD437). The synthetic-lethal and synthetic-viable interactions were analysed by comparing the CFAP20-KO line with the parental RPE1-iCas9 WT by first normalizing end-point reads based on t = 0 reads, as described previously50. We used an adapted version of DrugZ, termed IsogenicZ, which can be found at https://github.com/kdelint/IsogenicZ.
Immunostaining
Cells were grown on coverslips and fixed with 4% formaldehyde. By incubating with 0.5% Triton X-100 in PBS for 5 min, cells were permeabilized, followed by blocking with 100 mM glycine for 10 min. After washing with WB buffer (0.5% bovine serum albumin (BSA) and 0.05% Tween 20 in PBS), coverslips were incubated with the primary antibody (Supplementary Table 7) in WB buffer for two hours at room temperature. Cells were then washed extensively and labelled with their corresponding secondary antibody (Supplementary Table 7) in WB buffer containing 0.1 μg ml−1 DAPI for one hour at room temperature. Finally, the coverslips were washed extensively with PBS and mounted in Polymount (Brunschwig, 18606).
Immunostaining for detection of RNA–DNA hybrids
Indirect immunofluorescence with S9.6 antibody against RNA–DNA hybrids was performed as previously described57. Imaging of RNA–DNA hybrids using GFP–RNaseH1(D210N) was performed as described previously31.
Recovery of RNA synthesis
Cells were irradiated with UV-C light (12 J m−2), allowed to recover for the indicated periods and pulse-labelled with 400 μM 5-ethynyl-uridine (EU; Jena Bioscience) for one hour, followed by a 15 min medium-chase with DMEM without supplements. Cells were fixed with 3.7% formaldehyde in PBS for 15 min, permeabilized with 0.5% Triton X-100 in PBS for 10 min at room temperature and blocked in 1.5% BSA (Thermo Fisher Scientific) in PBS. Nascent RNA was visualized by click-iT chemistry, labelling the cells for one hour with a mix of 60 μM Atto azide–Alexa 594 (ATTO-TEC), 4 mM copper sulfate (Sigma), 10 mM ascorbic acid (Sigma) and 0.1 μg ml−1 DAPI in a 50 mM Tris-buffer (pH 8). Cells were washed extensively with PBS and mounted in Polymount (Brunschwig).
Microscopic analysis of fixed human cells
Images of fixed samples were acquired on a Zeiss AxioImager M2 wide-field fluorescence microscope equipped with 63× Plan-Apo (1.4 NA) oil-immersion objectives (Zeiss) and an HXP 120 metal-halide lamp was used for excitation. Fluorescent probes were detected using the following filters for DAPI (excitation filter, 350/50 nm; dichroic mirror, 400 nm; emission filter, 460/50 nm), Alexa 488 (excitation filter, 470/40 nm; dichroic mirror, 495 nm; emission filter, 525/50 nm) or Alexa 647 (excitation filter, 640/30 nm; dichroic mirror, 660 nm; emission filter, 690/50 nm). Images were recorded using ZEN 2012 (blue edition, v.1.1.0.0) and analysed in Image J (v.1.47–1.48). Graphs were plotted and analysed using GraphPad Prism 10 (10.2.3), Microsoft Excel 365 and Adobe Illustrator 2022, as described in Supplementary Table 9.
Quantitative image-based cytometry
Quantitative image-based cytometry was performed as described previously58. Colour-coded scatter plots and bar charts of asynchronous cell populations were generated with Spotfire data visualization software (v.10.10.1; TIBCO). Representative scatter plots and bar charts are shown.
Pairwise fluorescent competitive growth assay
Cell lines stably expressing either GFP or mCherry were seeded in a 1:1 ratio (30,000 cells per 6 wells). Cells were grown as usual and split every three days. During trypsinization, samples were taken at each time point. Cell pellets were washed with PBS followed by incubation in 2% formaldehyde in PBS for 15 min. Samples were quenched with glycine, washed with PBS, fixed in ice-cold methanol and stored at −20 °C. On the day of analysis, pellets were washed once with PBS and resuspended in 350 µl PBS. An AECE NovoCyte flow cytometer and NovoExpress software (Agilent) were used for analysis. For immunostaining, cells were grown simultaneously on coverslips and fixed in 4% formaldehyde at the corresponding time points. After permeabilization with 0.5% Triton X in PBS, cells were mounted with ProLong Gold Antifade Mountant with DNA Stain DAPI (Invitrogen, P36935).
Clonogenic growth assays
Cells were plated at low density in 6-cm culture dishes and allowed to attach, and were grown for ten days in growth medium supplemented with the indicated concentrations of the drugs. To visualize clones, cells were fixed with NaCl and stained with methylene blue. Formed clones were manually counted.
CellTiter-Glo assays
In a Costar black, clear-bottom 96-well plate, cells were seeded (WT and CFAP20-KO, 200 per well; BRCA1-KO, 400 per well) in medium containing increasing doses of olaparib or dimethyl sulfoxide (DMSO; 0.1% final DMSO concentration). Wells with no cells were included as a background luminescence control. After six days, the viability measurement was performed according to the manufacturer’s protocol. In brief, CellTiter-Glo substrate was dissolved in CellTiter-Glo buffer (Promega), and 100 µl of this was added to 100 µl fresh medium per well. The plate was briefly shaken and after equilibration, luminescence was recorded on a SpectraMax iD3 microplate reader (Molecular Devices). Luminescence values were corrected for background and for each cell line, normalized to wells treated with DMSO. Data were exported to GraphPad Prism 9.3.1 for further analysis.
DNA fibre spreading assay
Treatments with different compounds are shown in each experiment. Cells were labelled with 25 µM 5-chloro-2’-deoxyuridine (CldU) (Merck; Supplementary Table 2) for 20 min and washed three times with PBS, followed by labelling with 250 µM IdU (Merck; Supplementary Table 3) for 20 min. Labelled cells were collected and resuspended in 1× cold PBS. Two microlitres of the cell suspension was spotted on a positively charged slide (VWR) and then mixed with 7 µl of lysis buffer (200 mM Tris-HCl pH 7.4, 50 mM EDTA and 0.5% (w/v) SDS). The cells were incubated in lysis buffer horizontally for 5 min and then tilted at about 45°, allowing the drop to run by gravity. The DNA spreads were air-dried at room temperature and were then fixed in methanol/acetic acid (3:1) at room temperature for 10 min and stored at 4 °C overnight. Slides were processed as previously described59. Fibres were visualized and imaged using a Zeiss Axio Imager-M2 wide-field fluorescence microscope equipped with 40× Plan-Apo (1.4 NA) oil-immersion objectives (Zeiss) and an HXP 120 metal-halide lamp was used for excitation. Images were recorded and analysed with ZEN 2012 (blue edition, v.1.1.0.0) and analysed in Image J (v.1.53). Replication-fork speed (kb min−1) was calculated on the basis of the assumption that 1 µm of DNA fibre corresponds to 2.59 kb, as previously shown60.
DNA fibre assay with S1 nuclease
For the DNA fibre assay with the ssDNA nuclease (S1 nuclease), cells were labelled with 25 µM CldU for 15 min, washed three times with PBS and labelled again with 250 µM IdU for one hour. Cells were treated and processed as previously shown42,59.
scEdU–seq
The scEdU–seq procedure was performed according to a method described previously21. RPE1 WT and CFAP20-KO were labelled with 15-min pulses of EdU (10 μM). The cells were trypsinized, fixed in 70% ethanol and kept at −20 °C for 24 h. Then, the samples were resuspended and washed in 1 ml wash buffer (47.5 ml RNAse-free H2O, 1 ml 1 M HEPES pH 7.5, 1.5 ml 5 M NaCl, 3.6 µl pure spermidine solution, with an additional 0.05% Tween, and 4 µl ml−1 0.5 M EDTA). Next, biotin-PEG3-azide was conjugated to the EdU molecules through a CuAAC click reaction, followed by staining with DAPI. Single S-phase RPE1 cells were then sorted into 384-well plates for scEdU–seq processing. After sorting, libraries were prepared as follows: proteinase K digestion, NlaIII genome digestion, DNA blunt ending, A-tailing and adapter ligation incorporating cell barcodes and unique molecular identifiers (UMIs). Single-cell libraries were pooled and bound to MyOneC1 streptavidin beads to capture DNA replication fragments. These fragments were released by heat denaturation and filled in using the Klenow enzyme. The libraries underwent amplification through in vitro transcription, reverse transcription and PCR, followed by Illumina sequencing (NextSeq1000 P3 2×100 bp). The code for analysis and plotting can be accessed on GitHub21.
DRIP–qPCR
Approximately 1 ×107 cells per condition were lysed in 1.6 ml TE buffer supplemented with 82 μl of 10% SDS and 10 μl of 10 mg ml−1 proteinase K and incubated at 37 °C overnight. DNA was isolated by phenol:chloroform:isoamyl alcohol (25.24:1, v/v) extraction and isopropanol precipitation. DNA was reconstituted in 130 μl TE buffer, transferred to AFA microTUBEs with snap caps and sonicated for 4 min using a Covaris E220 sonicator (140 peak incident power, 10% duty factor and 200 bursts per cycle). Sonicated DNA was quantified on a NanoDrop 2000c spectrophotometer. For immunoprecipitation, 4 μg of DNA was resuspended in 150 μl 1× binding buffer (10 mM Na3PO4 pH 7, 140 mM NaCl and 0.05% Triton X-100), 10% removed as input DNA and the remaining sample bound to 6 μg of S9.6 antibody in 1× binding buffer overnight at 4 °C. Protein A/G agarose beads were added for two hours. Bound beads were washed three times in 1× binding buffer for 10 min at 4 °C. Elution was performed in elution buffer (50 mM Tris-HCl pH 8, 10 mM EDTA, 0.5% SDS and proteinase K) for 45 min at 55 °C with agitation. Eluted DNA was purified by phenol:chloroform:isoamyl alcohol (25.24:1, v/v) extraction and ethanol precipitation. Enrichment analysis of RNA–DNA hybrids in input and immunoprecipitation samples was performed by qPCR using the primers listed in Supplementary Table 6.
DRIP–seq
DRIP–seq was performed as previously described61 with minor modifications. Samples were sequenced using an Illumina NextSeq500 or HiSeq X, using paired-end sequencing with 42 bp or 151 bp from each end.
BrU–seq
Cells were grown to 80–90% confluency in three 15-cm plates per condition and incubated for 30 min with 2 mM BrU (Sigma, 850187). After incubation, cells were lysed in Trizol (Thermo Fisher Scientific, 15596018) and BrU-containing RNA was isolated as previously described62. cDNA libraries were made from the BrU-labelled RNA using the Illumina TruSeq library kit and paired-end 151-bp sequenced using the Illumina NovaSeq platform at the University of Michigan Advanced Genomics Core. Single-end or paired-end sequencing data were used for downstream analyses.
ChIP–seq
Cells were grown to 80–90% confluency and cross-linked with 0.5 mg ml−1 disuccinimidyl glutarate (Thermo Fisher Scientific) in PBS for 45 min at room temperature. Cells were washed once with PBS, followed by incubation with 1% formaldehyde for 20 min at room temperature. Fixation was stopped by adding glycine in PBS to a final concentration of 0.1 M for 3 min at room temperature. This was followed by washing with cold PBS and collection of the cells in 0.25% Triton X-100, 10 mM EDTA (pH 8.0), 0.5 mM EGTA (pH 8.0) and 20 mM HEPES (pH 7.6) in milli-Q. Chromatin was pelleted by centrifugation for 5 min at 400g and incubated in 150 mM NaCl, 1 mM EDTA (pH 8.0), 0.5 mM EGTA (pH 8.0) and 50 mM HEPES (pH 7.6) in milli-Q for 10 min at 4 °C. Chromatin was again pelleted by centrifugation and resuspended in ChIP buffer (0.15 % SDS, 1% Triton X-100, 150 mM NaCl, 1 mM EDTA (pH 8.0), 0.5 mM EGTA (pH 8.0) and 20 mM HEPES (pH 7.6) in milli-Q) to a final concentration of 15 × 106 cells per ml. Chromatin was sonicated to approximately one nucleosome using the Bioruptor Pico (Diagenode), with 8–15 cycles of 30 s on and 30 s off in a 4 °C water bath. RNAPII ChIP was performed using 28 µg of chromatin (22 µg for CCNC-KO and CFAP20/CCNC-dKO) + 40 ng of Drosophila spike-in chromatin (32.6 ng for CCNC-KO and CFAP20/CCNC-dKO; Active Motif, 53083) with 3 µl of RNAPII antibody and 1 µg spike-in antibody (Supplementary Table 6) by overnight incubation at 4 °C. TY ChIP was performed using 84 µg of WT chromatin and 60 µg of CFAP20-KO + TY-CFAP20 chromatin + 74 ng and 53 ng of Drosophila spike-in chromatin, respectively (Active Motif, 53083) with 5.7 µg of TY antibody (Diagenode, C15200054) and 1 μg spike-in antibody (Active Motif, 61686) (Supplementary Table 6) by overnight incubation at 4 °C. Protein–chromatin pull-down followed, with a 1:1 mix of protein A and protein G Dynabeads for RNAPII ChIPs, and protein A Dynabeads for TY ChIPs (Thermo Fisher Scientific, 10001D and 10003D). ChIP samples were washed extensively and purified using the QIAGEN MinElute kit. Sample libraries were prepared using the HiFi KAPA sample preparation kit and A–T-mediated ligation of NEXTflex adapters or xGen UDI-UMI adapters. Samples were sequenced using an Illumina NextSeq500 or HiSeq X, using paired-end sequencing with 42 bp or 151 bp from each end.
TTchem–seq
TTchem–seq was performed as described previously63. For TTchem experiments in WT or CFAP20-KO cells, this included depletion of rRNAs using the QIAseq FastSelect rRNA depletion kit (QIAGEN), followed by library preparation using the TruSeq Stranded Total RNA kit (Illumina, 20020596). For CFAP20-KO cells expressing WT GFP–CFAP20 or GFP–CFAP20(R100C), no ribosomal RNA was performed. The libraries were amplified according to the manufacturer’s instructions, pooled and paired-end sequenced on a DNBSEQ-G400 (BGI) system.
Definition of replication origins
OK–seq data in untreated RPE1 cells were downloaded from a previous report5 (datasets GSM3130725 and GSM3130726). Sequences were trimmed using TrimGalore (v.0.6.5) and aligned to hg38 using STAR (v.2.7.7a) with the genome file GCA_000001405.15_GRCh38. Duplicate reads were removed using SAMtools (v.1.11) with fixmate -m and markdup -r settings. Replication initiation zones were subsequently defined using the replication fork directionality analysis R toolkit (OKseqHMM v.2.0.0; available at https://github.com/CL-CHEN-Lab/OK-Seq; ref. 64), with read coverage threshold 6 for GSM3130725 and 1 for GSM3130726, and smoothing window size 15 kb. Initiation zones present in both datasets were identified using mergePeaks of HOMER tools (v.4.8.2)65, with -d given. Origins were defined as the centre of initiation zones. For all origins, their nearest TSS was defined using annotatePeaks of HOMER tools, together with the distance between the TSS and the origin. Here, a negative distance represents an origin upstream of the TSS (CD transcription relative to replication), whereas a positive distance represents an origin downstream of a TSS (HO transcription relative to replication). To allow for clean transcription versus replication analyses, we further selected only origins for which the nearest TSS was not preceded by another gene within 5 kb upstream of the TSS (Extended Data Fig. 2a). This resulted in a list of 2,040 origins.
ChIP–seq, DRIP–seq, BrdU–seq and TTchem–seq data analysis
For all sequencing data, a sequencing quality profile was generated using FastQC (v.0.11.9). Sequences were trimmed using TrimGalore (v.0.6.5). For ChIP–seq, reads were aligned to the human genome 38 GCA_000001405.15_GRCh38 and Drosophila genome BDGP6 using bwa-mem tools (BWA, v.0.7.17)66. For DRIP–seq, reads were aligned to the human genome 38 GCA_000001405.15_GRCh38 using bwa-mem tools (BWA, v.0.7.17)66. Only uniquely or primary mapping and high-quality reads (>q30) were included in the analyses. For BrU–seq and TTchem–seq, reads were aligned to hg38 using STAR (v.2.7.7a)67 with the genome file GCA_000001405.15_GRCh38. For ChIP–seq, BrU–seq and TTchem–seq data, duplicate reads were removed using SAMtools (v.1.11) with fixmate -m and markdup -r settings. Bam files were converted into stranded TagDirectories (with fixed fragment length 150–200 when automated fragment length definitions varied extensively) and UCSC genome tracks using HOMER tools (v.4.8.2)65. Example genome tracks were generated in IGV (v.2.4.3). A list of 2,040 origin coordinates was defined using data derived from a previous study5, as described in ‘Definition of replication origins’. A list of 49,948 gene coordinates was obtained from the UCSC genome database selecting the ‘knownCanonical’ table containing the canonical TSSs per gene68. To prevent contamination of binding profiles, genes were selected to be non-overlapping with at least 2 kb between genes and a minimal size of 3 kb (n = 9,944). From this, a set of 3,000 actively transcribed genes was selected by calculating gene-size-corrected read densities of BrU–seq data in WT cells, using the AnnotatePeaks.pl tool of HOMER with default settings. These 3,000 actively transcribed genes were used in downstream analyses, unless stated otherwise. For all DRIP–seq, ChIP–seq, BrU–seq and TTchem–seq experiments, read-density profiles around origin or TSS/TTS coordinates were defined using the AnnotatePeaks.pl tool of HOMER, using the default normalization to 10 million reads. For ChIP–seq experiments around transcribed genes, reads were normalized to the number of identified spike-in reads. Individual datasets were subsequently processed into heat maps or binding profiles using R (v.4.0.5) and Rstudio (v.1.1.423)69. Where indicated, average read-density profiles were generated after trimming 10% of the data (trim-mean 0.1; removing the top 5% and bottom 5% of datapoints) to remove extreme values.
Metaprofiles of TSSs in CD and HO orientations
We aligned TSSs with either a negative distance (CD) or a positive distance (HO) relative to the nearest origin. We subsequently generated average read-density profiles of RNAPII ChIP–seq, BrU–seq and DRIP–seq for all 1,395 CD and 408 HO genes at a maximum distance of 75 kb from the origin. We also sub-selected HO TSSs into those at 75–50 kb (n = 37), 50–25 kb (n = 80) or 25–0 kb (n = 291) upstream of the origin, and CD TSSs into those at 0–25 kb (n = 1,199), 25–50 kb (n = 143) or 50–75 kb (n = 53) downstream of the origin. These analyses provide a transcription-centred view of replication.
Statistics and reproducibility
Experimental data were plotted for statistical analysis in GraphPad Prism 10.2.3 (GraphPad). In figures showing all data points, each coloured circle represents a single cell, and the black circles represent the median of each independent biological repeat—of which there were at least two—as indicated for each experiment. More information on the n of each experiment is provided in the source data. Statistical analyses were performed on the median of each independent biological repeat per experiment using one-way ANOVA after Dunnett’s or Šidák’s correction where appropriate, unpaired two-tailed t-test or Fisher’s exact test, as indicated in the figure legends. All experiments were independently repeated at least twice, with similar results obtained. All micrographs are representative images of experiments that were performed at least twice, with similar results. In the figures, the notation NS = P > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001 is used, and precise P values are provided in the figure legends and the source data.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.