Cell lines
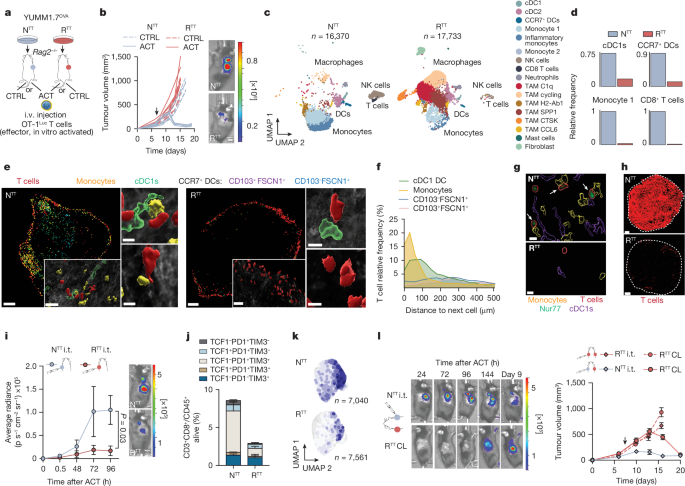
YUMM1.7 and YUMM3.3 mouse melanoma62 cell lines (obtained from M.âBosenberg, Yale University) were cultured in Dulbeccoâs modified Eagleâs medium (DMEM)âF12 produced in-house. A375, M249 (ref. 63) (obtained from J.âMassague, MSKCC), KPAR64 (obtained from J.âDownward, Francis Crick Institute) and EPP2 (ref. 65) (obtained from J.âZuber, IMP) cell lines were cultured in DMEM (Gibco). LOX48 (obtained from J.âMassague, MSKCC), CT-26 (ref. 66) and NCI-H358 cell lines were purchased from the American Type Culture Collection and cultured in RPMI-1640 (Gibco). The NCI-H358 RTT derivative was generated by culturing NCI-H358 parental cells in the presence of 1âμM KRAS inhibitor (Amgen) for 90âdays until cells became resistant. YUMM1.7OVA clones and all NTT and RTT derivatives were generated as previously described15. RTT BRAFi-resistant cancer cells (YUMM1.7 and YUMM3.3 model) and all genetically engineered derivatives were cultured continuously in 100ânM dabrafenib (Selleckchem). MEKi-resistant cancer cells were cultured continuously in 10ânM trametinib (Selleckchem). Human NTT and RTT melanoma cell line derivatives (A375, M249 and LOX) were generated as previously described48, and RTT cells were maintained in culture on 1âµM vemurafenib (LC-Labs). HEK-293T cells were purchased from Takara (Lenti-X 293T, 632180) and cultured in DMEM high-glucose produced in-house. BMDCs were cultured according to an adapted version of a previously described protocol67. In brief, for the first 6â7âdays, cells were cultured at a density of 1âÃâ106 cells per ml. On dayâ4, fresh medium was added to minimize cell death. After that, cells were either seeded for assays or counted and re-seeded at a density of 300,000 cells per ml. BMDCs were cultured in full Tâcell medium supplemented with 200ângâmlâ1 FLT3L-Ig (BioXcell) and 5ângâmlâ1 GM-CSF (in-house produced). Bone-marrow-derived Ly6C+ monocytes were cultured in DMEM medium (Gibco). Human MONO-MAC-1 (obtained from J.âZuber, IMP) and BLaER-1 (ref. 68) (obtained from M.âGaidt, IMP) cell lines were cultured in RPMI-1640 (Gibco). All media for cell lines were supplemented with 10% FBS, 2âmM l-glutamine (Gibco) and 100âIUâmlâ1 penicillinâstreptomycin (Thermo Fisher). BLaER-1 and NCI-H358 cells were additionally supplemented with 1à sodium pyruvate. CD8+ Tâcells were cultured in full Tâcell medium containing RPMI-1640 supplemented with 10% FBS, 2âmM l-glutamine and 100âIUâmlâ1 penicillinâstreptomycin, 1à sodium pyruvate (Gibco), 1à non-essential amino acids (Gibco), 20âmM HEPES (produced in-house) and 0.05âmM β-mercaptoethanol (Millipore). All cells were cultured at 37â°C and 5% CO2. Cells were routinely tested negative for mycoplasma contamination. STR Profiling was performed in-house for the YUMM1.7, YUMM3.3, EPP2 and KPAR cell lines. Moreover, sensitivity to MAPK inhibitors was confirmed for A375, M249 and LOX (BRAFi), CT-26 (MEKi) and for NCI-H358 (KRAS inhibitor).
Animal experiments and ethics
All mice were bred and housed in pathogen-free conditions with a housing temperature of 22â±â1â°C, 55â±â5% humidity and a photoperiod of 14âh of light and 10âh of dark. Within each experiment, age-matched and sex-matched groups were used. B6.129S(C)-Batf3tm1Kmm/J (Batf3â/â) mice, B6(Cg)-Zbtb46tm1(HBEGF)Mnz/J (zDC-DTR) mice, B6.Cg-Tg(Itgax-cre)1-1Reiz/J (Cd11c–cre) mice and NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice were purchased from Jackson Laboratories. B6.Cg-Rag2tm1.1Cgn/J Ly5.2 (Rag2â/â), BALB/c and C57BL/6J mice were obtained from the Vienna Biocenter in-house breeding facility. ItgaxcrePtger2â/âPtger4fl/fl mice were provided by J.âBoettcher (TUM, Munich). For Rag2â/âBatf3â/â strain generation, Batf3â/â mice were crossed to Rag2â/â mice, and homozygous offspring (Rag2â/ââÃâBatf3â/â) were confirmed by genotyping and used in subsequent experiments to evaluate the lack of cDC1s in the context of ACT. For Rag2â/â zDC-DTR strain generation, zDC-DTR mice were crossed to Rag2â/â mice and homozygous offspring were confirmed by genotyping and used in subsequent experiments to evaluate the effects of DC depletion. For ACT experiments and injection of YUMM1.7OVA cell lines, Rag2â/â mice were used. For the injection of YUMM3.3, KPAR and EPP2 cell lines, C57BL/6 mice were used. For the injection of the CT-26 cell line, BALB/c mice were used. For the generation of BMDCs and Ly6C+ monocytes, bones (femurs and tibias) were collected from in-house-bred C57BL/6 mice. For all above strains, mice were used between 6 and 12âweeks old. For OT-1Luc CD8+ Tâcell isolation, 6â24-week-old OT-1Luc Thy1.1 mice69 were used. All mouse experiments were performed according to our licence approved by the Austrian Ministry (GZ: MA58-2260492-2022-22; GZ: 340118/2017/25; BMBWF-66.015/0009-V/3b/2019; GZ: 801161/2018/17; and GZ: 2021-0.524.218 and their amendments). Mice were euthanized when the humane end point was reached (for example, weight lossâ>â20%, signs of distress and pain), when tumours displayed signs of continuous necrosis or when tumours reached the maximum allowed tumour volume of 1,500âmm3.
Tumour cell injections
For subcutaneous injections, mice were anaesthetized with 2â4% isoflurane. For the YUMM1.7OVA model and all its derivatives, 0.5â1âÃâ106 YUMM1.7OVA cancer cells were subcutaneously injected into the flank of each mouse in a volume of 50âµl. For contralateral experiments, alternating flanks were used for the injection of NTT and RTT cells to avoid preferential growth biases. For the YUMM3.3 model, 0.3â1âÃâ106 cells were subcutaneously injected in a volume of 50âµl. For the CT-26 model, 0.25âÃâ106 cells were subcutaneously injected in 50âµl. For the KPAR model 0.35âÃâ106 cells were subcutaneously injected in 50âµl. For the EPP2Luc cell line derivative, orthotopic injections were performed as previously described65. In brief, surgeries were performed under isoflurane (2â4%) anaesthesia on a heated plate. A small incision on the upper left quadrant of the shaved abdomen was made and the was spleen identified. After externalization of the pancreas, 1âÃâ106 cells were intrapancreatically injected. Organs were re-situated, and the peritoneum closed with a resorbable 6-0 Vicryl suture, followed by skin closure with sterile wound clips. Animals received intraperitoneal (i.p.) injections of 5âmgâkgâ1 carprofen pre-emptively and every 12â48âh after surgery. The health status of mice was monitored daily, and the tumour burden was assessed by BLI. All cell lines were resuspended in PBS mixed 1:1 with Matrigel (Corning) in the final injection volume. Subcutaneous tumours were monitored by calliper measurements every 2â4âdays, and tumour volume was calculated according to the following formula: volumeâ=â(DâÃâd2)/2, in which D and d are the long and short tumour diameters, respectively.
Isolation and activation of naive OT-1Luc CD8+ Tâcells
Spleen and lymph nodes were isolated from OT-1Luc mice, and red blood cell lysis was performed with ammoniumâchlorideâpotassium lysis buffer (Thermo Fisher) according to the manufacturerâs protocol. Tâcell isolation was performed using a Magnisort mouse CD8+ naive Tâcell enrichment kit (Thermo Fisher) according to the manufacturerâs protocol. Tâcells were activated for the first 24âh by seeding them on a plate coated with 2âµgâmlâ1 anti-CD3 (145-2C11, eBioscience) overnight, and adding 1âµgâmlâ1 anti-CD28 (37.51, eBioscience) and 20ângâmlâ1 carrier-free IL-2 (BioLegend). Tâcells were expanded for approximately 6â7âdays in the presence of IL-2 and maintained daily at a concentration of 1âÃâ106 cells per ml in fresh Tâcell medium.
ACT, intratumoral injection and BLI
Unless otherwise specified, when tumours reached a volume of 100â150âmm3, 4âÃâ106 in vitro-activated OT-1Luc CD8+ Tâcells were i.v. injected into mice in a volume of 100âµl PBS. For i.t. injections, 4âÃâ106 in vitro-activated OT-1Luc CD8+ Tâcells were injected in a volume of 50âµl PBS. For measuring Tâcell infiltration by BLI, d-luciferin (150âmgâkgâ1, Goldbio) was injected retro-orbitally or by tail vein injection into anaesthetized mice, and mice were imaged with an IVIS machine (Caliper Life Sciences) and analysed using Living Image software (v.4.4; Caliper Life Sciences). In NTT tumours, Tâcell recruitment to the tumour is detectable by BLI within 24â48âh. This initial recruitment is followed by a phase of Tâcell expansion, with peak BLI signals between 96 and 120âh. Hence, we depict 96âh post-ACT images (unless otherwise specified in figure legends) as a suitable time point to assess Tâcell expansion in immune-permissive TMEs.
In vivo treatments
For treatment with ICB, mice were i.p. injected with anti-PD1 (clone RMP1-14, BioXcell) and anti-CTLA4 (clone 9D9, BioXcell) in 100âµl of PBS when tumours reached a volume of 150â200âmm3 (usually between 6 and 8âdays after injection). The YUMM3.3 model was treated with 200âµg anti-PD1/anti-CTLA4, the CT-26 model with 100âµg anti-PD1, and the EPP2 model with 100âµg anti-PD1. ICB treatment was administered every 3âdays and continued for at least for 3âweeks, as indicated in the figure legends. Control mice were treated with an isotype control antibody (rat IgG2a anti-trinitrophenol, clone 2A3, BioXcell, and mouse IgG2b, clone MPC-11, BioXcell). For COX2i treatment, celecoxib (LC Laboratories) was reconstituted in a 60:40 (DMSO to PEG400, dH2O) mixture as previously described53. Etoricoxib (Sellekchem) was dissolved first in a small volume of DMSO and then in 1% sodium carboxymethyl cellulose. COX2i was given by oral gavage every day (30âmgâkgâ1) in a volume of 200âµl. For both COX2i regiments (celecoxib and etoricoxib), the treatment was started at dayâ3 after injection, when tumours were palpable, and continued every day until the termination of the experiment. 5-AZA (Sigma-Aldrich) was reconstituted in DMSO to a stock concentration of 10âmgâmlâ1 and further diluted in PBS for in vivo treatments and given as i.p. injections (1âmgâkgâ1) in 100â250âµl every 3âdays, as previously described54. For NK cell depletion, 200âµg anti-NK1.1 (clone PK136, BioXcell) was administered every 3âdays through i.p. injections, starting at day 1 after tumor induction. NK cell depletion was confirmed by flow cytometry. For blocking Tâcell egress from the lymph node, mice were given an i.p. injection of 20âµg per mouse of FTY720 (Sigma) in 100âµl saline. Treatment was started on the day of Tâcell transfer and administered for 5â7 consecutive days. Control mice received saline injection. FLT3L (recombinant FLT3L-Ig, hum/hum, BioXCell) treatment (30âµg per mouse in 100âµl PBS i.p.) was started at dayâ3 after injection and administered every day for 9 consecutive days. In vivo IFNAR blockade was performed with InVivoMab anti-mouse IFNAR-1 (clone MAR1-5A3, BioXcell) and was administered i.p. (200âµg per mouse) in 100âµl. For IFNγ, the neutralizing anti-mouse IFNγ monoclonal antibody was used (clone XMG1.2, BioXcell). Treatment was started on the day of tumour engraftment and administered every 3âdays. InVivoMab IgG1 isotype control (BioXCell) was used as the control. For experiments in which CD8 depletion was performed, mice were treated with 50âµg anti-CD8 (clone 2.43, in-house produced), whereas control mice were treated with isotype control (rat IgG2b anti-keyhole limpet haemocyanin, clone LTF-2) starting the day before tumour engraftment and then every 3âdays.
DC vaccination with BMDCs
BMDCs were cultured with FLT3L and GM-CSF as described above. Atâdayâ10â12 after isolation, DCs were activated overnight with polyI:C (5âµgâmlâ1, Invitrogen), pulsed with recombinant SIINFEKL peptide (5âµgâmlâ1, Genscript) and sorted by FACS on the basis of alive MHCII+CD103+CD11c+ cells. Next, 1âÃâ106 cells in a volume of 50âµl PBS were i.t. injected. Control mice received 50âµl PBS. For DC vaccinations, 2 doses of i.t. injections were administered on dayâ4 and dayâ6 after tumour engraftment.
In vivo depletion of DCs with diphtheria toxin
For generation of bone marrow chimeras, Rag2â/â Ly5.1 mice were preconditioned (2Ã5âGy), before transferring back 10âÃâ106 bone marrow cells by i.v. injection. As donor mice, Rag2â/â Ly5.2 zDC-DTR mice were used. After 8âweeks of reconstitution, mice were used for experiments. NTT cells were injected, and DCs were depleted by injecting 25âµgâkgâ1 of body weight of diphtheria toxin (Sigma-Aldrich) i.p. in PBS, starting on the day of tumour engraftment and then every 3âdays for 3â4âdoses. Reconstitution efficiency and depletion of intratumoral DCs was confirmed by flow cytometry.
Lentivirus generation and cell transduction
Lenti-X (HEK-293T) cells were transfected with 4,000âng of the plasmid of interest, 2,000âng of VSV-G plasmid and 1,000âng of PAX2 plasmid using polyethylenimine (Avantor). Virus-containing supernatant was collected 24âh and 48âh after transfection and subsequently filtered through a 0.45âµm filter. The cell lines of interest were transduced with the collected virus mixed with 8âµgâmlâ1 polybrene (Merck).
Generation of CRISPRâCas9 KO and overexpression cell lines
Doxycycline-inducible Cas9 (iCas9) clones from parental cell lines were generated to allow inducible expression of Cas9. sgRNAs were chosen on the basis of the best VBC score70 (Supplementary Table 7) and were cloned into a vector containing a puromycin selection marker and mCherry or eGFP (hU6-sgRNAâPuroRâmCherry/eGFP). sgRNAs targeting the ROSA26 locus were used as controls for KO cell lines. After transduction, cells were selected with puromycin (5â8âµgâmlâ1) for 5âdays. All sgRNA sequences are provided in Supplementary Table 7. For the generation of single-cell-derived clonal cell lines, cells were FACS sorted on the basis of the fluorescent marker on the sgRNA backbone, at 1 cell per well into 96-well plates. To avoid immunogenicity caused by antibiotic selection markers or fluorophores in the YUMM3.3 model, we transiently transfected the cell lines with an all-in-one vector containing Cas9, the sgRNA of interest and eGFP (U6-IT-EF1As-Cas9-P2A-eGFP). For transient transfection, 7,000âng of the plasmid with polyethylenimine was used, and single-cell clones were established. For IRF3/7 overexpression, synthesized cDNA sequences were ordered from Twist Biosciences and cloned into two different expression vectors with distinctive selection/fluorescent markers (SFFV-IRF3âmCherry and SFFV-IRF7âPuroR). After transduction, cells were selected with puromycin (5â8âµgâmlâ1 for 5âdays) and bulk FACS-sorted on the basis of mCherry expression. The same cell line engineered with an empty vector containing an mCherry and a puromycin resistance cassette was used as a control. KO and overexpression of the target proteins was confirmed by genotyping, western blotting or quantitative PCR with reverse transcription (RTâqPCR). For the YUMM1.7 and YUMM3.3 Ptgs2 KO cell lines, single-cell-derived clonal cell lines were generated, and several were tested in vivo for growth kinetics.
EP2 and EP4 KO in Tâcells
sgRNAs targeting the Ptger2 and Ptger4 mouse genes were designed according to the VBC score70 and cloned into a dual hU6-sgRNA-mU6-sgRNA-EF1α-mCherry-PuroR backbone (Supplementary Table 7). As a control, we used a sgRNA targeting a gene desert in chromosomeâ1. The lentiviral vector was produced as described above. Tâcells were isolated from Cas9âOT-1 mice, which were a gift from J.âZuber (IMP), as described above. Twelve hours after CD3/CD28 activation, Tâcells were spin-infected with the lentiviral vector containing the sgRNAs in a 1:1 ratio for 1âh at 32â°C and 800g. At 12âh after infection, Tâcells were removed from the activation plate, washed with PBS and cultured in the presence of 20ângâmlâ1 IL-2. Selection with puromycin was performed 30âh after viral transduction. Before ACT, mCherry levels were assessed, and KO was confirmed by functional in vitro assays.
Flow cytometry and cell sorting
For flow-cytometry-based characterization of the TME, tumours were isolated between dayâ7 and 11 after injection, cut into pieces and digested for 1.5âh at 37â°C with collagenaseâA (1âmgâmlâ1, Roche) and DNAse (20âµgâmlâ1, Worthington) in unsupplemented RPMI-1640 medium. Digested tumours were strained through a 70âµm filter and resuspended in FACS buffer (0.5% BSA and 2âmM EDTA in PBS). Fc-block was performed with anti-CD16/32 (clone 2.4G2, Pharmingen) for 10âmin at 4â°C to avoid Fc-specific antibody capture, and staining for cell surface markers was performed for 30âmin at 4â°C. For intracellular staining, a Foxp3 Transcription Factor staining kit was used (eBioscience). Live/dead exclusion was performed by staining with the fixable viability dye eFluor780 (1:1,000, eBioscience). DCs were defined in most experiments as MHCII+CD11c+CD24+ out of alive CD45+ cells. cDC1s were identified as CD103+CD11bâ out of the total DCs, cDC2s as CD103âCD11b+ and inflammatory cDC2 as CD103âCD11b+AXL+. AXL was previously described to identify inflammatory cDC2s37. Monocytes were defined as Ly6C+CD11b+F4/80â, and inflammatory monocytes were identified as monocytes that were Ly6A+. Ly6A was previously described to identify monocytes expressing high levels of ISGs38. Macrophages were defined as Ly6CâF4/80+Cd11b+. Acquisition of the samples was performed using a BD LSR Fortessa machine (BD Biosciences) with FACS Diva software (v.9.0.1), and analysis was conducted using FlowJo software (v.10.8 or newer). For cell sorting, a BD Aria cell sorter (BD Biosciences) with FACS Diva software (v.9.0.1) was used.
Antibodies for flow cytometry
The following antibodies (all anti-mouse) were used for flow cytometry stainings (target (clone, catalogue number, manufacturer, dilution)): AXL PE-Cy7 (MAXL8DS, 25-1084-82, eBioscience, 1:200); CD103 PerCP/cyanine5.5 (2E7, 121415, BioLegend, 1:100); CD103 PE (2E7, BioLegend, 121405, 1:100); CD11b APC (M1/70,17-0112-81, eBioscience, 1:200); CD11b PerCP/cyanine5.5 (M1/70, 101229, BioLegend, 1:200); CD11c BV605 (HL3, 563057, BD Pharmigen, 1:100); CD11c FITC (N418, 117305, BioLegend, 1:100); CD24 BV510 (M1/69, 101831, BioLegend, 1:100); CD24 FITC (M1/69, 11-0242-82, eBioscience, 1:100); CD279/PD-1 BV785 (29F.1A12, 135225, BioLegend, 1:200); CD279/PD-1 FITC (29F.1A12, 135213, BioLegend, 1:200); CD40 APC (3/23, 124611, BioLegend, 1:200); CD45 BV711 (30-F11, 103147, BioLegend, 1:500); CD45 FITC (30-F11, 103107, BioLegend, 1:500); CD86 BV510 (GL-1, 105039, BioLegend, 1:100); CD3 BV605 (17A2, 564009, BD Horizon, 1:100); CD3 AF647 (17A2, 100209, BD Horizon, 1:100); CD3 AF488 (17A2, 100212, BD Horizon, 1:100); CD8a eFluor 450 (53-6.7, 48-0081-80, eBioscience, 1:100); CD8a AF647 (53-6.7, 128041, BioLegend, 1:100); MHCI (H-2Kb) APC (AF6-88.5.5.3, 17-5958-82, Bioscience, 1:200); MHCI (H-2Kb) PE (AF6-88.5.5.3, 17-5958-80, Bioscience, 1:200); MHCII (I-A/I-E) eFluor450 (M5/114.15.2, 48-5321-80, eBioscience, 1:200); MHCII (I-A/I-E) APC (M5/114.15.2, 107613, BioLegend, 1:200); NK-1.1 BV711 (PK136, 108745, BioLegend, 1:100); TCF1 PE (S33-966, 564217, BD Pharmigen, 1:50); TIM3 BV711 (RMT3-23, 119727, BioLegend, 1:100); CD88 PE (20/70, 135805, BioLegend, 1;100); Ly-6A/E (Sca-1) FITC (D7, 108105, BioLegend, 1:100); SIINFEKL-HK2B PE (25-D1.16, 12-5743-81, Invitrogen, 1:100); F4/80 PE (BM8, B123110, BioLegend, 1:200); and rat IgG1, K Isotype control PE (R3-34, 5546, BD Pharmigen). Further information is provided in Supplementary Table 8.
RNA extraction of cancer cells sorted from tumours, in vitro cell lines and myeloid cells
Tumours were surgically removed between daysâ10 and 12 after injection. The tissue was processed as described above, and cancer cells were isolated by flow cytometry on the basis of alive, CD45â cells and a fluorescent marker. For cancer cell lines and in vitro assays with myeloid cells, cells were washed with PBS and snap-frozen in liquid nitrogen and kept at â70â°C until further processing. RNA was extracted using a magnetic bead-based RNA extraction protocol (in-house produced). In brief, cells were lysed and incubated with beads together with DNaseâI (NEB) followed by magnetic isolation. RNA was purified by further elution with nuclease-free water.
RTâqPCR
Reverse transcription was performed for cDNA formation with 1âµg of RNA per sample utilizing a LunaScript RT SuperMix kit (NEB) according to the manufacturerâs instructions. RTâqPCR was performed with 10âng cDNA per sample either with Luna Universal qPCR master mix (NEB) or an in-house produced MTD qPCR Dye 2à HS master mix according to the manufacturerâs protocol. Each sample included four technical replicates. The RTâqPCR reaction was carried out in a Bio-Rad CFX384 realâtime cycler and contained 1âmin of initial denaturation (95â°C) and 45 annealing cycles lasting 15âs at 95â°C and 30âs at 60â°C. The analysis of gene expression levels was determined by the quantification cycles (Cq). Internal controls and the housekeeping gene GAPDH were used to correct for differences in sample quality and to normalize expression values. qPCR primer pair sequences are listed in Supplementary Table 7.
In vitro assays with BMDCs
For cancer cell CM experiments, supernatants (in full Tâcell medium) from confluent cancer cells were collected after 48âh, filtered through a 45âµm filter and frozen at â70â°C until further use. Full Tâcell medium was supplemented with 20âµM of the COX1/2i indomethacin (Selleckchem) or 5ânM of the MEKi trametinib (Selleckchem) for the evaluation of MAPK and COX1/2 activity before media conditioning. BMDCs were differentiated as described above and collected at dayâ6. Next, 0.5â1âÃâ106 cells were seeded in triplicate in a 12-well plate in CM and treated with 10âµgâmlâ1 InVivoMab anti-mouse IFNAR-1 antibody or InVivoMab IgG1 isotype control (BioXCell). Cells were cultured for 24âh, collected and processed for flow cytometry analysis or RNA extraction. For treatment with PGE2 and IFNβ, cells were collected at dayâ6 and seeded at a concentration of 0.5â1âÃâ106 cells per ml. Cells were treated for 24â48âh with recombinant PGE2 (100ângâmlâ1, Sigma-Aldrich) and recombinant mouse/human IFNβ (R&D Systems) at the concentrations indicated in the corresponding figures. Same volumes of acetone and PBS were used as a control for PGE2 and IFNβ, respectively.
Isolation of bone-marrow-derived Ly6C+ monocytes for intratumoral injection and in vitro assays
Ly6C+ monocytes were directly isolated from the bone marrow of CD45.1+ C57BL/6 mice using a monocyte isolation kit (Miltenyi Biotec) following the manufacturerâs instructions. For intratumoral monocyte transfer, 1âÃâ106 monocytes were i.t. injected into NTT and RTT tumours established in CD45.2+ Rag2â/â mice. Tumours were isolated for FACS analysis 72âh after intratumoral transfer. For in vitro assays to assess effects of PGE2, Ly6C+ monocytes were seeded at a density of 1âÃâ106 cells per ml and cultured in recombinant IL-4 and GM-CSF (both produced in-house) and exposed to 200ângâmlâ1 PGE2 or vehicle for 3 or 5âdays. For CM experiments, monocytes were seeded at a density of 1âÃâ106 cells per ml in CM obtained from NTT, RTT or RTT IRF3/7 cells with or without 20âµM COX1/2i (indomethacin) during media conditioning and subsequently supplemented with or without 10âµgâmlâ1 InVivoMab anti-mouse IFNAR1 anti-mouse (BioXCell) or isotype IgG1 control (BioXCell).
In vitro monocyte co-culture assay
Ly6C+Ly6A+ or Ly6C+Ly6Aâ monocytes were FACS-sorted from NTT tumours grown in Rag2â/â mice or BALB/c mice and co-cultured for 72âh with naive OT-1 T cells (1:3 ratio: 100,000 monocytes for 300,000 naive OT-1 cells) previously labelled with 0.25âµM CFSE for 30âmin at 37â°C.
In vitro human monocyte assays
BLaER-1 cells were transdifferentiated into monocytes as previously described68. In brief, BlaER-1 transdifferentiation medium was freshly prepared by adding 10ângâmlâ1 human recombinant (hr-)IL-3 (PeproTech), 10ângâmlâ1 hr-M-CSF (PeproTech) and 100ânM β-oestradiol (Sigma-Aldrich) to complete RPMI medium. Cells were resuspended in transdifferentiation medium and plated in a 12-well plate at 0.7âÃâ106 cells per ml. Cells were incubated at 37â°C for 5â6âdays until mature monocytes were differentiated. For CM experiments, BLaER-1 or MONO-MAC-1 human monocytes were seeded at a density of 0.7âÃâ106 cells per ml in CM obtained from NTT or RTT cells from the human melanoma cell lines A375, M249 and LOX or the human NSCLC cell line NCI-H358 with or without 20âµM COX1/2i (indomethacin) during media conditioning. Cells were cultured in CM for 24âh and collected for RNA extraction, as described above.
Evaluation of pMHCI cross-dressing on monocytes
For mismatched MHCI haplotype experiments, 1âÃâ106 YUMM1.7OVA NTT cells from C57BL/6 origin (H-2Kb) were injected in the flank of BALB/c (H2-Kd) mice. BALB/c mice were treated with anti-CD8 (50âµg in 100âµl, in-house produced), whereas control mice were treated with isotype control (rat IgG2b anti-keyhole limpet haemocyanin, clone LTF-2) starting the day before tumour engraftment and then every 3âdays to avoid Tâcell-mediated mismatched MHCI rejection of YUMM1.7 cells. On dayâ10, tumours were collected and processed for flow cytometry staining of H2-Kb or FACS-sorted on the basis of Ly6A expression for in vitro assays.
Sample preparation for scRNA-seq
For scRNA-seq experiments involving TME characterization, tumours were isolated at dayâ10 after injection (72âh after ACT) and were processed as described above. The CD45+ live fraction was isolated by FACS, and approximately 1âÃâ105 cells were collected. For scRNA-seq of OT-1 Tâcells, tumours were isolated 5âdays after i.t. injection of 4âÃâ106 Tâcells. Alive Tâcells were isolated from tumours by FACS for CD45+CD3+CD8+ markers. Dissociated cell concentrations were measured using NucleoCounter NC250 (Chemometec) following the manufacturerâs instructions. For scRNA-seq samples from experimentsâ3 and 4 (see below), a Chromium Next GEM Single Cell Fixed RNA Sample preparation kit was used according to the manufacturerâs protocol. In brief, 1âÃâ106 cells were fixed for 22âh at 4â°C, quenched and long-term stored at â80â°C according to 10x Genomics Fixation of Cells & Nuclei for Chromium Fixed RNA profiling (CG000478) using a Chromium Next GEM Single Cell Fixed RNA Sample preparation kit (PN-1000414, 10x Genomics). About 250,000 cells per sample were used for probe hybridization using a Chromium Fixed RNA Kit, Mouse Transcriptome, 4rxn à 4BC (PN-1000496, 10x Genomics), pooled equally and washed following the Pooled Wash Workflow as described in the Chromium Fixed RNA Profiling Reagent kit protocol (CG000527, 10x Genomics). For all the other scRNA-seq samples, a Chromium Next GEM Single cell 3â² kit with Dual Index was used according to the manufacturerâs instructions. GEMs were generated on Chromium X (10x Genomics) with a target of 10,000 cells recovered, and libraries prepared according to the manufacturerâs instructions (CG000527, 10x Genomics). Sequencing was performed on NovaSeq S4 lane PE150 (Illumina) with a target of 15,000 reads per cell.
scRNA-seq analysis of CD45+ TME
CD45+ immune cells were collected in four different 10x Genomics sequencing experiments. Experimentâ1, Chromium Single Cell 3â² scRNA-seq samples were pre-processed using cellranger count (v.6.1.1) (YUMM3.3 samples: NTT/108155 and RTT/108157). Experimentâ2, 3â² CellPlex multiplex experiment with 4 samples pre-processed using cellranger multi (v.6.1.1) (YUMM1.7OVA samples: NTTâ+âACT, RTTâ+âACT, RTT Ptgs1/2 KOâ+âACT, RTT CTRL ROSA26â+âACT). Experiments 3 and 4, Chromium Flex multiplex experiments with 4 samples each pre-processed using cellranger multi (v.7.1.0) and the built-in Probe Set (v.1.0.1 mm10-2020-A). Experimentâ3, YUMM1.7OVA samples: RTT mCherry CTRL, RTT IRF3/7, RTT COX2i and RTT COX2iâ+â5-AZA, all ACT treated. Experimentâ4, YUMM1.7OVA contained biological replicates of experimentâ2 samples and untreated YUMM1.7OVA samples (noA): NTTnoA/271221, RTTnoA/271222, NTT/271223 ACT, RTT/271224 ACT. The prebuilt 10x Genomics mm10 reference refdata-gex-mm10-2020-A was used. Further processing was performed in R (v.4.2.2) with Seurat (v.4.3.0). For generating a CD45+ immune reference map, we integrated cells from the first three experiments as follows. The cellranger filtered featureâbarcode matrices were used, retaining cells with more than 1,000 detected genes and less than 15% of mitochondrial and less than 40% of ribosomal RNA reads. An integrated featureâbarcode matrix from the three experimental batches was generated accounting for the inclusion of a probe-based assay by keeping genes found in at least five cells in each experiment and excluding ribosomal and mitochondrial genes. Data were log-normalized, scaled (regressing out the difference between the G2M and Sâphase signature scores), dimensionality reduction was performed using principal component analysis on the top 3,000 most variable genes, and batch correction across batches was performed using Harmony71 (v.0.1.1). The 40 harmony embeddings were used for UMAP visualizations. The first 40 harmony dimensions were used to identify immune cell subclusters with a resolution of 0.5 that were further assigned to cell types using known markers and publicly available myeloid reference datasets21,72. Cells were scored for the expression of published signatures using the AddModuleScore function73. Wilcoxon rank-sum test implemented in Presto (v.1.0.0) was used to identify differentially expressed genes (DEGs). Seuratâs reference-based mapping was used to predict cell-type identity and map cells of the biological replicate experiment to our annotated reference set using the FindTransferAnchors and MapQuery functions after a quality control process retaining cells between 1,000 and 4,500 detected genes for 27,1222 and 27,1224 cells, respectively, and 1,300 and 8,000 detected genes for 27,1221 and 27,1223 cells, respectively, and limiting count tables to the gene universe of the reference. Depth-normalized counts for pseudobulk and GSEA functional analyses of this experiment were generated using cellranger aggr. Differences between ACT and untreated conditions (no ACT) from the replicate experiment (experimentâ4) were explored on a pseudo-bulk level in an unsupervised clustering analysis with heatmap visualization. The fibroblast cluster was removed before further processing. Sum aggregation on the depth-normalized UMI counts was followed by variance stabilizing transformation, selection of the 300 most variable genes, standardization, k-means clustering (kâ=â3) and Enrichr analysis against the Reactome_2022 using Enrichr. The relative frequency bar plots depict the changes in the relative abundance of a cell type across different experimental conditions. For each condition, we calculated the normalized abundance of a specific cell type by comparing the absolute number of the cell type to the absolute number of all cells in the same condition. This normalization accounts for differences in total number of cells captured between conditions. We then calculated the relative cell abundance of the cell type in all conditions of the experiment. This was done by comparing the normalized abundance of the cell type to the sum of normalized abundances of the same cell type across conditions of the experiment. This step produces values between 0 and 1 for each condition for each cell type, with the sum of these values across all conditions of the experiment equalling 1 for each cell type.
scRNA-seq analysis intratumoral CD8+ OT-1Luc T cells
Single-cell gene expression of isolated NTT and RTT T cells was assayed in a Chromium Flex experiment, and read processing was performed using cellranger multi (v.7.1.0) using probeset (v.1.0.1 mm10-2020-A). Cellranger-filtered featureâbarcode matrices were used and further filtered to retain cells with more than 800 detected genes, less than 10% of mitochondrial and less than 10% of ribosomal RNAs reads, and removal of cells of contaminant clusters was identified using SingleR and ImmGen reference (fibroblasts, MoMac populations). Data were log-normalized and scaled, and dimensionality reduction was performed using principal component analysis on the top 2,000 most variable genes. Harmony was used for the integration of cells from different samples, and 15 harmony embeddings were used for UMAP visualizations. Published tumour single-cell data were used for signature scoring29. Gene lists are provided in Supplementary Table 2.
RNA velocity analysis
To understand differentiation trajectories of myeloid cells within the TME, we performed RNA velocity analysis74 of the MoMac compartment. Loom files containing the splicing annotation were created for each sample using the velocyto run command from the package velocyto (0.17,17) with default parameters and with no masked intervals. The loom files were combined with the scRNA-seq object that had been filtered to keep the data for monocyte and macrophage populations (Monocyte_1, Monocyte_2, Infl_Mono, TAM_CCL6, TAM_Ctsk, TAM_C1q, TAM_H2-Ab1, TAM_Spp1 and TAM_cycling) and for each condition (NTT, RTT and RTT Ptgs1/2 KO). First-order and second-order moments were computed using scvelo (0.2.5) pp.moments (n_pcs = 30, n_neighbors = 30), and the dynamical model was run with default parameters. Python (v.3.8.12) was used.
SCENIC analysis
Gene regulatory networks for each cell population in each condition were calculated using SCENIC75. The motif database used was mm9-tss-centered-10kb-7species.mc9nr.feather. The co-expression network was calculated using GENIE3. The gene regulatory network was built using SCENIC wrapper functions.
Analysis of publicly available myeloid datasets and inflammatory signatures
For the melanoma and lung samples from a previously published23 dataset (Gene Expression Omnibus (GEO) identifier GSE154763), the raw counts were pre-processed as described in the publication, and clustering was calculated using a resolution of 0.8. The monocyte and inflammatory monocyte gene set was derived by using the wilcoxauc() function from presto and by selecting the genes with a log fold changeâ>â0.6 (Supplementary Table 2). Then, the gene symbols were converted to human symbols. The human inflammatory monocyte gene set was used to calculate an enrichment score per cluster. In brief, the gene average expression was calculated for each cluster in the LUNG and MEL datasets on normalized data. Then, an enrichment score was calculated using GSVA with the following parameters: minSizeâ=â5, maxSizeâ=â500, kcdfâ=ââGaussianâ. The projection of the signature on the UMAP embedding was done using the function AddModuleScore() and then plotting the resulting score using FeaturePlot() with min.cutoffâ=â0.3 for the inflammatory monocyte score and min.cutoffâ=â0.4 for the Monocyte_1 score. For another dataset43, the annotated seurat-object for myeloid populations corresponding to the original figure 4a was obtained, and gene sets were analysed as described above. For querying published inflammatory gene signatures, a previously published ISG+ DC signature37 was generated by taking the top DEGs in the cDC2 cluster37. The Bosteels Inf-cDC2 DC signature was previously generated37 and was obtained by re-analysing the scRNA-seq dataset (GEO identifier GSM4505993), in which the top 20 DEGs were taken in the identified inflammatory cDC2 cluster. All of them were subsequently scored in our dataset using the AddModuleScore73, and the resulting score was plotted using FeaturePlot(). Gene lists are provided in Supplementary Table 2.
Single-cell spatial transcriptomics of human melanoma samples
Single-cell spatial transcriptomics profiling was performed using the CosMx technology (Nanostring). Biopsy samples were obtained from patients with an age at diagnosis that ranged from 24 to 85âyears with a median of 66âyears; 34% were women and 66% were men. We obtained cell-segmented data for 74 FOVs (an area of 500âÃâ500âµm) from tissue microarray cores of 34 melanoma metastases, in total consisting of 980 genesâÃâ171,536 cells. Tumour samples were obtained from 21 lymph nodes, 7 subcutaneous metastases, 1 lung metastasis and 1 brain metastasis and 4 not annotated, from 31 patients containing 72 FOVs. Two FOVs were from tonsils as control. Most tumour tissue were from patients who were treatment-naive at the time of surgery. Tissue collection was approved by the Regional Ethics committee at Lund University (numbers 191/2007 and 101/2013). Patients provided informed consent. The majority of tissue microarray cores contained tertiary lymphoid structures, and FOVs were preferentially directed to these regions. Low-quality FOVs, cells with <20 counts and potential multiplets of cells (area exceeding the sample geometric meanâ+â5 standard deviation) were discarded. Using Seurat, genes for which the mean expression was below 3à the median of the negative probe mean expression, and genes with the highest 99% quantile expression, MALAT1 and IGKC (due to potential spillover to neighbouring cells), were removed, which retained 641 genes. The data were normalized using SCTransform76, counts that were zero before SCTransform were restored, and counts were log-transformed as log2(counts+1). The top 30 principal components were used for UMAP reduction and clustering (k.paramâ=â15, resolutionâ=â0.5, Louvain algorithm). Resulting clusters were assigned to biological annotations using known marker genes, and annotations were mapped back to FOV coordinates. Expression of C1QC, CXCL9 or CXCL10â>â0 was considered as positive. Cell-type fractions were derived for each FOV. Pearson correlation values between cell-type fractions across FOVs were determined and displayed. In Fig. 2i, CXCL9+CXCL10+ macrophage/DCs (numberâ9 and numberâ10) were either CXCL9+ or CXCL10+. Macrophage/DCs (numberâ5) were negative for CXCL9, CXCL10 and C1qC.
Generation of the TME-COX and TME-IRF3/7 signature
For the TME-COX signature, the FindMarkers function was used in Seurat, with tresh.useâ=â0.25 and min.pctâ=â0.1, to compare RTT CTRL (ROSA26) and RTT Ptgs1/2 KO scRNA-seq samples. The top DEGs (log2 fold change ⤠1.5, adjusted Pâvalueâ<â0.05) were used and converted to human orthologues using DIOPT77. For the TME-IRF3/7 signature, the FindMarkers function was used in Seurat, comparing RTT CTRL (mCherry) and RTT IRF3/7 and taking the top 40 DEGs.
TME signatures in immunotherapy-treated human samples
Gene expression data for patients receiving ICB were obtained from a previous study49 (NCBI BioProject accession number PRJEB23709). The TME-COX, TME-IRF3/7 and CD8+ Tâcell scores for each tumour sample were defined as the geometric mean of the expression values of each of the gene sets, respectively (Supplementary Table 4). The univariate Cox proportional hazards models, in which the TME-COX and TME-IRF3/7 scores were included as continuous variables, were used for testing the statistical association between gene signature expression and patient survival, separately for both signatures. The tumour samples were then divided into three groups on the basis of the signature score (bottom third, mid-third and top third) and KaplanâMeier plots were generated for visualization. The association between signature expression and CD8+ Tâcell abundance was evaluated by calculating the Personâs correlation coefficient between the signature score and a CD8+ score for each signature separately. For this, all scores were normalized to a median of zero and standard deviation of one. The two overlapping genes were removed from the CD8+ signature before comparing it to TME-IRF3/7 signature expression. For evaluating the enrichment of TME-COX and TME-IRF37 gene signatures in responder and non-responder patients to TIL therapy (baseline) from a previous study43, mouse gene identifiers were first converted to human orthologues (with DIOPT v.9; best dcoreâ=âyes, best score reverseâ=âyes, DIOPT scoreâ>â7) and single-cell level signature enrichment scores for the âhumanizedâ gene sets were calculated using AddModuleScore_UCell78.
Analysis of transcriptomics data
For plots shown in Fig. 3a, a cut-off of adjusted Pâvalueâ<â0.05 and log2 fold changeâ>â2 and <ââ2 was used on DEGs expressed in YUMM1.7OVA NTT and RTT GFP+ cancer cells FACS-sorted out of tumours (Supplementary Table 3). Pathway enrichment analysis was performed using Enrichr79,80. For the plot in Extended Data Fig. 5i, upstream regulator analysis (Ingenuity)81 was used to identify upstream regulators using DEGs with an adjusted Pâvalueâ<â0.05.
Quantification of PGE2 and IFNβ by ELISA
For in vitro analysis of PGE2 production, 2âÃâ106 cells were seeded in 10âml medium, and supernatants were collected after 48âh and kept at â70â°C until analysis. For IFNβ, 0.3âÃâ106 cells were seeded, and 1âml of supernatant was collected from confluent cells in a 6-well plate after 48âh of culture and kept at â70â°C until analysis. For analysis of PGE2 and IFNβ from mouse tumours, whole tumours were isolated between daysâ4 and 10 after engraftment, accurately weighed and immediately snap-frozen in liquid nitrogen. They were stored at â70â°C until further processing. For PGE2 analysis, tumours were subsequently digested using a MACS dissociator according to the manufacturerâs protocol in PBS supplemented with 1âmM EDTA and 10âµM indomethacin. Lysate was further diluted in dissociation buffer depending on the tumour condition and weight (100âµl per mg of tumour) and further quantified using a PGE2 ELISA kit (Cayman) or a mouse IFNβ Quantikine ELISA kit (Biotechne) according to the manufacturerâs protocol. Values were normalized by taking into account dilution factors and tumour weight. For human IFNβ analysis from human cells, 1âÃâ106 A375, M249, LOX or NCI-H358 cells were injected into NSG mice and collected on dayâ21. Tumours were processed as described above and quantified using a Human IFNβ Quantikine ELISA kit (Biotechne) according to the manufacturerâs protocol.
Eicosanoid analysis from tumours by HPLCâMS
YUMM1.7OVA NTT and RTT tumours were isolated at dayâ10 after injection and weighed, and a solution of isopropanol and methanol (1:1, v/v) was added to the tissue for metabolite extraction. The material was subsequently homogenized and incubated for 1âh at â20â°C. The samples were then centrifuged at 14,000g for 3âmin. A second extraction round was performed by adding 80% methanol and H2O (v/v) to the pellet and centrifuged, and both supernatants were combined. Finally, the samples were incubated for another 2âh at â20â°C, and after final centrifugation, the supernatants were stored at â70â°C until further analysis. Samples were subsequently measured on a ZIC-pHILIC column or a RP column. Metabolites were annotated using the compound discoverer 3.0 software (Thermo Fisher) using an internal database or the mzCloud database (at least 75% match on the basis of measured molecular weight and MS2 spectra). For filtering, a RSD of corrected quality control areas was used, being less than or equal to 25%. Group CV of at least 1 group is less than or equal to 40%.
Western blotting
Cells were lysed with RIPA buffer (Cell Signaling Technology) supplemented with complete Protease Inhibitor Cocktail (Sigma Aldrich) and HALT phosphatase inhibitor (Thermo Fisher Scientific). Lysates were sonicated and cleared by centrifugation at 14,000g for 10âmin at 4â°C. Protein concentrations were quantified according to the manufacturerâs instructions using a BCA Protein Assay kit (Pierce, Thermo Fisher Scientific). Immunoblotting was conducted according to standard protocols. The primary antibodies used for immunoblotting were as follows: anti-vinculin (Sigma-Aldrich, 1:1,000), anti-COX2 (CST,1:1,000) and anti-H3 (acetyl K27) (Abcam, 1:5,000). The secondary antibodies used were as follows: anti-rabbit IgG HRP-linked (Cell Signaling Technology, 1:10,000) and anti-mouse IgG HRP-linked (Cell Signaling Technology, 1:10,000).
Volumetric IF microscopy and image analysis
Volumetric microscopy of mouse tumours was performed as previously described9. In brief, tumours were fixed in Antigenfix solution (Diapath) for 6â8âh, dehydrated in 30% sucrose overnight, embedded in TissueTek OCT freezing medium (Sakura Finetek) and stored at â80â°C. Using a Leica CM3050âS cryostat, consecutive sections of 50âµm thickness were generated, subsequently permeabilized, blocked and stained in 0.1âM Tris (Carl Roth) supplemented with 1% BSA, 0.3% Triton X-100 (Merck), normal mouse serum (Merck) and donkey serum (Merck). Stained sections were mounted in Mowiol (Merck) and imaged on an inverted TCS SP8 confocal microscope (Leica) using a HC PL APO CS2 Ã20/0.75 NA objective. Images were acquired as tiled image stacks, covering whole tumour sections in the xy plane, with 2âµm z-spacing to provide 3D image volumes of at least 20âµm depth. For further analyses, images were adaptively deconvoluted using the Leica TCS SP8 LIGHTNING tool (v.3.5.7.23225) and analysed using Imaris 9.9 software (Oxford Instruments). The Imaris surface generation tool was used to reconstruct and visualize 3D objects for individual cells. Where indicated, signals outside rendered cells were masked to visualize intracellular proteins. For analysis of immune cell infiltration by histocytometry, statistics for object localizations were exported into Excel (v.16.88; Microsoft) and analysed using GraphPad Prism software (GraphPad). Quantification of the number of cells was performed relative to the volume of the imaged section. Interacting cells were described as being less than <5âµm apart from each other.
Antibodies for immunofluorescence microscopy
The following antibodies were used for staining of mouse tissues: anti-CD3 (BioLegend, clone 17A2), anti-CD103 (R&D Systems, goat polyclonal), anti-FSCN1 (Santa Cruz Biotechnology, clone 55-k2), anti-Ly6C (BioLegend, clone HK1.4) and anti-MHCII I-A/I-E (BioLegend, clone M5/114.15.2). All antibodies were either validated by the manufacturer or were previously reported for IF microscopy. The populations were defined as follows: Tâcells (CD3+), monocytes (Ly6C+CD103âMHCII+), cDC1s (FSCN1âCD103+MHCII+), CCR7+ cDC1 (FSCN1+CD103+MHCII+) and CCR7+ cDC2 (FSCN1+CD103âMHCII+). Nur77âGFP was directly assessed by transferring Nur77âGFP reporter OT-1 T cells.
Meta-analysis of NSAID immunotherapy cohorts
The meta-analysis was performed in accordance with the updated Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) reporting guidelines82. The literature search was conducted using the PubMed (MEDLINE) database and last updated on 31 December 2023. The full search strategy is available in Supplementary Table 6. The literature review included studies of (1) adult patients with (2) melanoma or NSCLC (3) undergoing FDA-approved immunotherapy, including anti-PD1, anti-PD-L1 or anti-CTLA4, (4) co-medication with NSAIDs and (5) available sufficient patientsâ outcome data to calculate odds ratios for overall response rates or hazard ratios for progression-free and overall survival. Patients were not excluded when receiving concomitant chemotherapy and/or radiotherapy. Included studies report time of overall survival, time of progressions-free survival and overall response rates (defined as complete responses and partial responses divided by patient population). All studies published since 1 January 2011 (FDA approval of first immunotherapy, for example, ipilimumab) were included. Survival data are reported as univariate or multivariate hazard ratios; if both were available, multivariate analysis was prioritized. Odds ratios and hazard ratios with 95% CIs for overall response rates, progression-free and overall survival from included studies were utilized to calculate the pooled odds and hazard ratios. The heterogeneity of the pooled results was evaluated using Q-tests to assess between-study heterogeneity and quantified by the Higgins I2 test. If P was <0.10 for the Q-test or I2 was >50%, significant heterogeneity was assumed, and the random-effects model was used to summarize the data. Statistical analysis was performed using R software (v.4.3.2) with meta (General Package for Meta-Analysis, v.7.0-0).
Statistical analysis and reproducibility
Statistical analyses were performed using GraphPad Prism (v.9.1.2 or newer) and Microsoft Excel (v.16.88). Normality of the data distribution was calculated using a DâAgostino and Pearson test or ShapiroâWilk test. The number of samples (n) used per experiment and the statistical test used are indicated in the figure legends. All in vitro and in vivo experiments were repeated at least twice and always with multiple replicates, except for the following experiments that were performed only once: scRNA-seq involving pharmacological treatment of the YUMM1.7 RTT model, intratumorally injected Tâcells and the YUMM3.3 model. IF stainings for which representative images are shown were repeated at least twice, except for the NTT in Batf3â/â and Nur77 reporter experiment, which was performed once but with nâ=â3 tumours and was also confirmed with flow cytometry. Pharmacological combination treatments of the KPAR model were performed once. No statistical methods were used to determine sample size for in vivo experiments, and numbers were chosen on the basis of previous preliminary experiments. Scientists were not blinded to experimental groups, and experiments were repeated by different investigators. Mice were randomly assigned to treatment groups on the basis of tumour size at the day of treatment start or randomly allocated across separate cages when treatment had to be started at dayâ3. Pâvaluesâ<â0.05 were considered significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.