Mice and reagents
All animal experiments were conducted in accordance with UK Home Office guidelines and were approved by the University of Cambridge Animal Welfare and Ethics Review Board. Littermate controls or age- and sex-matched mice were used in experiments as indicated. All mice, unless otherwise stated, were housed at the University of Cambridge University Biomedical Services Gurdon Facility or the Babraham Institute Biological Support Unit. Wild-type C57BL/6 mice were obtained from Charles River. Ptprca (CD45.1) congenic, OT-1 TCRtg, Rag2–/– and MMTV-PyMT (B6.FVB-Tg(MMTV-PyVT)634Mul/LellJ) mice were obtained from the Jackson Laboratory. Arhgef1-KO (Arhgef1Tm1a) mice have previously been described12. Tbxas1-KO (Tbxas1tm1Swl) and Tbxa2R-KO (Tbxa2rtm1Cof) mice, as previously described44,49, were provided by S.-W. Lin. Arhgef1fl/fl mice were generated by crossing Arhgef1Tm1a mice with FlpO-deleter mice50, and subsequently crossed with Cre-expressing strains, Ncr1cre (Ncr1tm1.1(icre)Viv), Lyz2cre (Lyz2tm1(cre)Ifo) and Cd4cre (Tg(Cd4-cre)1Cwi) mice to generate conditional-knockout mice, respectively17,18,19. Mice were genotyped by Transnetyx. Pf4cre Ptgs1fl/fl mice have been described previously42 and were housed at the animal facility of the “G. d’Annunzio” University of Chieti-Pescara and the animal experiments were performed under the European Communities Council (EEC) Directive of 22 September 2010 (2010/63/EU) and the National Ethical Committee (authorization no. 434/2024-PR).
Tumour metastasis model
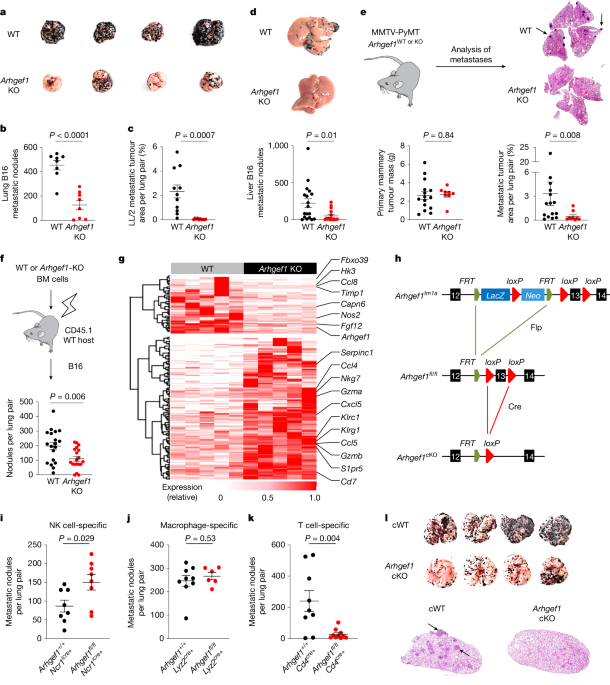
Mouse metastatic melanoma cell lines B16-F10 (purchased from Kerafast) and B78ChOva-mCherry (provided by M. F. Krummel)51 were passaged in DMEM (Thermo Fisher Scientific) supplemented with 10% FBS and antibiotics. For lung metastasis model: mice were injected intravenously with 5 × 105 B16-F10 cells or B78ChOva-mCherry cells in 150 µl Dulbecco’s PBS (DPBS) and lungs were dissected between day 14 and 17 for metastasis enumeration and subsequent analysis. LL/2 murine Lewis lung carcinoma cells were passaged in DMEM supplemented with 10% FBS and antibiotics. Mice were injected with 1 × 106 LL/2 cells in 150 μl DPBS and lungs were dissected on day 17. Lungs were fixed in 4% formalin before being embedded in paraffin. Sections were taken from the centre of the lungs and stained with H&E. Slide images were taken using a Pannoramic digital slide scanner (3DHistech) and analysed using QuPath Software. Tumour burden was calculated as a percentage of total tissue area for each sample. For the liver metastasis model: mice were anaesthetized with isoflurane and a small incision was made in the left flank to expose the spleen. 3.5 × 105 B16-F10 cells suspended in 50 μl DPBS were injected into the spleen. The wound was closed with sutures and skin staples. Mice were euthanized and livers were dissected on day 11 for metastasis enumeration and subsequent analyses. Aspirin (Aspégic, Sanofi Aventis and Sigma) was resuspended in drinking water at 600 mg l−1. TXA2 analogue U46619 (Cayman) was diluted in DMSO and delivered in drinking water at 50 μg kg−1. All the drinking water contained 1% sucrose and was replaced 3–4 times per week. For platelet depletion, mice were administered R300 antibody (Emfret) intraperitoneally at 0.25 mg kg−1 every 2–3 days from 1 day before intravenous injection of tumour cells. For inhibition studies, COX-1 inhibitor SC-560 (SelleckChem and Abcam), COX-2 inhibitor Celecoxib (SelleckChem), P2Y12 inhibitor Ticagrelor (SelleckChem) or vehicle control were administered at 30 mg kg−1 via daily oral gavage 5 days before intravenous injection of tumour cells and continuing throughout the study.
Spontaneous germline MMTV-PyMT cancer metastasis model
Mice were palpated for mammary tumours once per week. Mammary tumours were assessed by taking length and width measurements with digital callipers three times a week after the first palpable mammary tumour was detected. When the total mammary tumour area exceeded 2.25 cm2, mice were euthanized and lungs were collected and fixed in 4% formalin before being embedded in paraffin. Sections were taken from the centre of the lungs and stained with H&E. Slide images were taken using a Pannoramic digital slide scanner (3DHistech) and analysed using QuPath Software. Tumour burden was calculated as a percentage of total tissue area for each sample.
MC38 subcutaneous tumour model
MC38 mouse colorectal cancer cells were passaged in DMEM supplemented with 10% FBS and antibiotics. Mice were injected subcutaneously into one flank with 2.5 × 105 cells in 100 µl PBS. Tumour growth was assessed by taking length and width measurements with digital calipers 3 times a week starting from day 7 after injection.
L. monocytogenes infection and kinetic analysis
For experiments assessing the response of CD8+ T cells to bacterial infections, bacteria were grown in BHI medium to an OD600 of 0.1 before each experiment. The mice were infected with a sub-lethal dose of 5 × 106 colony-forming units attenuated (ΔactA) L. monocytogenes expressing OVA by intravenous administration52. Blood samples were collected via the tail vein at serial time points following infection. CD8+ T cell responses were detected and their phenotypes were analysed by flow cytometry.
Bone marrow reconstitution experiments
For bone marrow reconstitution experiments, C57BL/6 mice were administered 1,000 Gy total-body γ-radiation from a 137Cs source before intravenous injection of bone marrow cells from single-cell bone marrow preparations from 8- to 12-week-old mice. Mice were administered neomycin for two weeks following irradiation and reconstitution to limit infection risk, and were used in flow cytometry and cancer metastasis experiments at two to three months after reconstitution.
Flow cytometry
Single-cell suspensions from lymphoid tissues were prepared by mechanical dissociation through 40-µm cell strainers (BD Biosciences). Lungs were minced in media containing 20 µg ml−1 DNase I (Roche) and 1 mg ml−1 collagenase (Sigma Aldrich) and incubated with agitation at 37 °C for 40 min before also being dissociated through 40-µm cell strainers. Erythrocytes were lysed using ice-cold ACK Lysing Buffer (Gibco) for 5 min. Cells requiring intracellular staining of cytokines were stimulated prior to flow cytometry analysis using PMA, ionomycin, brefeldin A and monensin for 4 h in complete RPMI 1640 (Thermo Fisher Scientific). Viable cells were discriminated by first staining alone with Zombie UV fixable viability dye (Biolegend) or eFluor 780 fixable viability dye (eBioscience) in PBS, according to the manufacturer’s instructions. Cells were then incubated with specific surface antibodies on ice for 40 min in FACS buffer, in the presence of 2.4G2 monoclonal antibodies to block FcγR binding. Cell surface phosphatidylserine was labelled using the eBioscience Annexin V Apoptosis Detection Set (Thermo Fisher Scientific) according to the manufacturer’s protocol. For intracellular staining, the eBioscience Foxp3/Transcription Factor Staining Buffer Set (Thermo Fisher Scientific) or BD Cytofix/Cytoperm Fixation/Permeabilization Kit was used in accordance with the manufacturer’s instructions followed by intracellular staining with fluorochrome-conjugated antibodies for 40 min. Samples were acquired using Cytek Aurora (Cytek), BD LSR Fortessa (BD Biosciences) or Beckman CytoFLEX (Beckman Coulter) cytometers with their respective software: SpectroFlo (v3.3.0), BD FACSDiva (v8.0.1), and CytExpert (v2.5.0.77). Data were analysed using FlowJo v10.10.0 software (TreeStar LLC).
Antibodies used for flow cytometry are as follows: anti-CD103-Pacific Blue (2E 7, BioLegend, 121418, 1/200), anti-CD127-BV650 (A7R34, BioLegend, 135043, 1/200), anti-CD127-PE-Cy7-A7R34 (BioLegend, 135014, 1/200), anti-CD16/CD32 (93, BioLegend, 101302, 1/200), anti-CD25-PE-Cy7 (PC61, Invitrogen, 25-0251-82, 1/200), anti-CD25-BUV395 (PC61, BD, 564022, 1/200), anti-CD39-AF647 (Duha59, BioLegend, 143808, 1/400), anti-CD4-AF700 (RM4-5, BioLegend, 100536, 1/200), anti-CD4-BV650 (RM4-5, BioLegend, 100546, 1/400), anti-CD44-BV510 (IM7, BioLegend, 103044, 1/200), anti-CD44-BV786 (IM7, BD, 563736, 1/400), anti-CD44-PerCP-Cy5.5 (IM7, Invitrogen, 45-0441-82, 1/400), anti-CD44-APC (IM7, Invitrogen, 17-0441-83, 1/400), anti-CD45.2-ef506 (104, Invitrogen, 69-0454-82, 1/100), anti-CD45.2-FITC (104, BioLegend, 109805, 1/200), anti-CD61-PE (2C9.G3, Invitrogen, 12-0611-82, 1/200), anti-CD62L-BUV737 (MEL-14, BD, 612833, 1/400), anti-CD62L-APC (MEL-14, BioLegend, 104412, 1/400), anti-CD69-PECy5 (H1.2F3, BioLegend, 15-0691-82, 1/300), anti-CD69-PE-Dazzle (H1.2F3, BioLegend, 104536, 1/200), anti-CD8α-BUV395 (53-6.7, BD Horizon, 563786, 1/200), anti-CD8α-BUV805 (53-6.7, BD, 612898, 1/200), anti-CD8α-BV510 (53-6.7, BioLegend, 100752, 1/200), anti-CD8α-FITC, (53-6.7, Invitrogen, 11-0081-86, 1/200), anti-CD90.1-FITC (OX-7, BioLegend, 202503,1/100), anti-CD90.1-PerCP (OX-7, BD, 557266, 1/100), anti-Foxp3-APC (FJK-16S, eBioscience, 17-5773-82, 1/200), anti-IFNγ-BUV737 (XMG1.2, BD, 612769, 1/400), anti-IFNγ-FITC (XMG1.2, BioLegend, 505806, 1/200), anti-IL-2-PE (JES6-5H4, BioLegend, 503808, 1/200), anti-Ki67-PerCP-ef710 (SolA15, Invitrogen, 46-5698-80, 1/200), anti-KLRG1-APC (2F1/KLRG1, BioLegend, 138412, 1/200), anti-KLRG1-BV605(2F1/KLRG1, BioLegend, 138419, 1/200), anti-Ly108-APC (330-AJ, BioLegend, 134610, 1/200), anti-Ly6G-FITC (RB6-8C5, eBioscience, 11-5931-85, 1/400), anti-PD-1-APCef780 (J43, Invitrogen, 47-9985-82, 1/200), anti-PD-1-PE-Cy7 (RMP1-30, BioLegend, 109110, 1/200), anti-pErk T202/Y204-AF488, (197G2, Cell Signalling, 13214S, 1/100), anti-pErk T202/Y204-AF647 (197G2, Cell Signalling, 13148S, 1/100), anti-pS6 S235/6-PE (D57.2.2E, Cell Signalling, 5316S, 1/150), anti-ST2-PerCP-ef710 (RMST2-2, eBioscience, 46-9335-82, 1/200), anti-TCF-1-AF488 (C63D9, Cell Signalling, 6444S, 1/200), anti-TCRβ-BV570, (H57-597, BioLegend, 109231, 1/200), anti-TCRβ-FITC (H57-597, BioLegend, 109206, 1/200), anti-TCRβ-PerCP-Cy5.5 (H57-597, BioLegend, 109228, 1/200), anti-TIGIT-PE-Dazzle (1G9, BioLegend, 142110, 1/100), anti-TIGIT-PE (GIGD7, Invitrogen, 12-9501-82, 1/100), anti-TIM3-BV421 (RMT3-23, BioLegend, 119723, 1/100), anti-TIM3-BV785 (RMT3-23, BioLegend, 119725, 1/100), anti-TNF-APC (MP6-XT22, BioLegend, 506308, 1/200), anti-TNF-BV650 (MP6-XT22, BioLegend, 506333, 1/200), anti-TOX-PE (REA473, Miltenyi, 130-120-716, 1/200) and TER119-FITC (TER119, Invitrogen, MA5-17822, 1/200)
Computational analysis of flow cytometry data
Data were processed as previously described53,54. In brief, Flow Cytometry Standard (FCS) 3.0 files were first imported in FlowJo version 10.10.0 to eliminate dead cells by manual gating, and select CD45+ leukocytes, subjected to biexponential transformation, then exported for computational analysis by a custom-made script making use of PhenoGraph (K value set at 30). Here, we modified the Linux-community and the core.py script to fix the seed to “123456” (run in Python version 3.7.3). Data were then converted in comma-separated value (CSV) files and merged into a single file by using the pandas package. The obtained data, exported as new CSV file (one for each cluster), were further imported in FlowJo and analysed to define the percentage of cells positive for each protein as well as their median fluorescent intensity. Data were finally metaclustered using the gplots R package. UMAP was performed with the UMAP Python package.
Fluorescence-activated cell sorting
Pre-enrichment of CD8+ T cells from single-cell suspensions was done using the MagniSort Mouse CD8+ T cell Enrichment Kit (Thermo Fisher Scientific) according to the manufacturer’s protocol. Markers required for cell sorting were stained using flow cytometry cell surface antibodies, diluted 1/100, while cell suspensions were being labelled with the Enrichment Antibody Cocktail from the kit. Cells were filtered again and resuspended in RPMI 1640 containing DAPI for live/dead discrimination before sorting. Cell sorting was performed using a BD Fusion or Aria instrument (BD Biosciences). Cells were sorted into solutions of RPMI 1640 supplemented with 25% Charcoal-Stripped FBS (Gibco) before being prepared for experiments as described below.
Primary T cell culture
Naive CD8+ T cells were isolated by FACS from the spleens and lymph nodes of wild-type and Arhgef1-KO mice and stimulated with immobilized Ultra-LEAF purified anti-mouse CD3ε (BioLegend) and Ultra-LEAF purified anti-mouse CD28 (37.51; BioLegend) in culture medium containing rhIL-2 (5 ng ml−1; Peprotech) for 4–5 days. In some cases, cells were labelled with 2.5 μM CTV proliferation dye (Thermo Fisher) before stimulation, according to the manufacturer’s instructions.
Gα12/13 GPCR ligand stimulation assays
Naive CD8+ T cells were purified as described above by FACS from the spleens and lymph nodes of wild-type and Arhgef1-KO mice. Purified naive CD8+ T cells (0.8 × 105) labelled with 2.5 μM CTV were activated by plate-bound anti-CD3ε and soluble anti-CD28 (5 µg ml−1 each; BioLegend) in lipid-free medium containing 10% charcoal-stripped FBS for 5 days in the presence of rhIL-2 (5 ng ml−1) and Gα12/13 GPCR ligands as follows: CXCL12 (1 μg ml−1; R&D), Thrombin (10 units ml−1; Sigma Aldrich), PAR1 peptide (10 μM; Abcam), PAR2 peptide (10 μM; Abcam), S1P (5 μM; Acros Organics), 20:4 LysoPI (1 μM; Avanti Polar Lipids), 18:1 LysoPS (1 μM; Avanti Polar Lipids), LPA (1 μM; Avanti Polar Lipids), 9-HODE (1 μM; Cayman), histamine dihydrochloride (5 mM; Sigma Aldrich), prostaglandin E2 (0.5 μM; Cayman) and U46619 (5 μM; Calbiochem). At the end of the cell culture, cells were collected, and cell proliferation and activation were measured by flow cytometry. For the inhibition study, cells were treated with the TP inhibitor SQ 29548 (Cayman) at the concentration of 10 μM, beginning 1 h before stimulation and continuing until the end of the cell culture.
Acute T cell stimulation and analysis of protein phosphorylation
CD8+ T cells from wild-type and Arhgef1-KO OT-1 TCRtg mice were stimulated in vitro with 10 nM OVA257–264 peptide in the presence of rhIL-2 for 2 days, followed by a 3-day expansion with rhIL-2. Before TCR crosslinking, cells were serum-starved overnight in RPMI 1640 containing 0.1% fatty acid-free BSA (Sigma Aldrich). Cells were then incubated with soluble anti-CD3ε antibody (Thermo Fisher Scientific) at 4 °C for 20 min. After washing, CD3 molecules bound with anti-CD3ε antibodies were crosslinked using goat anti-Armenian hamster IgG (H+L) (Jackson ImmunoResearch) in a lipid-free medium containing 5 μM U46619 or DMSO control for 5 min. For western blots, following crosslinking, cells were immediately lysed in Pierce RIPA buffer with cOmplete Mini protease and PhosSTOP phosphatase inhibitor cocktails (Roche). Cells were then denatured in 2× Laemmli buffer (Bio-Rad) with 2-mercaptoethanol and boiled for 10 min at 98 °C before gel loading. Western blotting on PVDF membrane was performed using TGX reagents (Bio-Rad Laboratories) and protocols. Following transfer, blots were blocked with 5% BSA then incubated with primary antibodies against pAkt-Ser 473 (193H12, Cell Signaling), pan-Akt (9272, Cell Signaling), pS6-Ser 235/236 (D57.2.2E, Cell Signaling), S6 (5G10, Cell Signaling), pMEK1/2-Ser 217/221 (9121, Cell Signaling), MEK1/2 (9122, Cell Signaling), pERK1/2-Thr202/Tyr204 (D13.14.4E, Cell Signaling), ERK1/2 (3A7, Cell Signaling), ARHGEF1 (D25D2, Cell Signaling), β-actin (AC74, Sigma Aldrich) and GAPDH (1E6D9, Proteintech) with appropriate horseradish peroxidase-conjugated secondary antibodies (Bio-Rad). Horseradish peroxidase-conjugated secondary antibodies blots were developed using chemiluminescence (Thermo Fisher) and gel images were captured in the darkroom using photographic films. For inhibition studies, cells were pre-treated for 1 h with the following inhibitors: AKT inhibitor VIII (1 μM, Calbiochem), TP inhibitor SQ 29548 (10 μM, Cayman), ROCK inhibitors Y-27632 (30 μM, Tocris) and GSK269962A (10 μM, ApexBio) or PTEN inhibitor bpV(pic) (2.5 μM, Sigma Aldrich) before TCR crosslinking.
Ex vivo phosflow assay
Single-cell suspensions from the spleens and lymph nodes and lungs from mice of indicated genotype and treatments were prepared as described above except for using lipid-free medium throughout the procedure. Two-to-three million cells were incubated with specific surface antibodies and soluble anti-CD3ε antibody (Thermo Fisher Scientific) at 4 °C for 30 min. After washing, CD3 molecules bound with anti-CD3ε antibodies were crosslinked using goat anti-Armenian hamster IgG (H+L) (Jackson ImmunoResearch) in a lipid-free medium containing 5 μM U46619 or vehicle control for 5 min. Following crosslinking, the cells were immediately fixed with ice-cold 2% formaldehyde at 4 °C for 20 min. After fixation, the cells were permeabilized with ice-cold 90% methanol at 4 °C for 20 min. Phosphorylation of S6 and ERK was measured by flow cytometry using anti-pERK T202/Y204 AF488- or AF647-conjugated antibodies (197G2) and anti-pS6 S235/6-PE or -AF647 (D57.2.2E) antibodies.
Assays for quantification of RHOA activation
The activity of RHOA was measured using the RHOA Pull-Down Activation Assay Biochem Kit (Cytoskeleton). Following overnight serum starvation, 10 × 106 wild-type and Arhgef1-KO OT-1 TCRtg CD8+ T cells were stimulated with 5 μM U46619 for 5 min at 37 °C. Cells were immediately spun down, washed and lysed in the lysis buffer with the phosphatase and protease inhibitor cocktails at 4 °C. Fifty micrograms of Rhotekin-RBD beads (Cytoskeleton) were added to the lysates and rotated for 1 h at 4 °C. The samples were spun down, washed three times, denatured in 2× Laemmli buffer with 2-mercaptoethanol and boiled for 10 min at 98 °C before gel loading. The primary antibody against RHOA (Abcam) was used for western blotting.
Plasmids, cloning, retroviral transduction and CRISPR–Cas9 mutagenesis
Platinum-E ecotropic packaging cells (Cell Biolabs) were plated one day before transfections on poly-d-lysine-coated 10-cm plates (Corning) at a concentration of 6 × 106 cells per plate. Packaging cells were transfected with 10 μg of retroviral plasmid DNA encoding MSCV-IRES-Thy1.1 (pMIT) or pMIT-RHOACA along with 6 μg pCL-Eco plasmid DNA using Transit293 (Sigma Aldrich) in OptiMEM (Thermo Fisher Scientific) for 8 h in antibiotic-free medium. Medium was replaced 8 h after transfection and cells were incubated for a further 48 h. Retroviral supernatants were used to spinfect Cas9-expressing OT-1 TCRtg CD8+ T cells following stimulation with 10 nM OVA257–264 peptide for 24 h. In brief, CD8+ T cells were collected, suspended in viral supernatant and spun at 2,000g for 2 h at 32 °C in 24-well plates in the presence of 8 μg ml−1 polybrene (Sigma Aldrich) with slow acceleration and deceleration. Cells were cultured for 3–4 further days in rhIL-2-containing media prior to acute restimulation and phospho-flow assays.
For CRISPR–Cas9 mutagenesis, three different pairs of sgRNAs for Rhoa were designed using the CRISPick tool (Broad Institute), subcloned into optimized retroviral MSCV-sgRNA-puro-Thy1.1 vectors and retrovirally transduced to Cas9-expressing CD8+ T cells as described above. Deletion of Tbxa2r in naive T cells was as previously described55. In brief, three different sgRNAs targeting Tbxa2r: 5′-CCAGAGAAGCTCATGACAGG-3′, 5′-UUAGGAGCCAUGUGGCCCAA-3′ and 5′-CGAGGUGCCAUUGGGCCACA-3′ or their negative control: scrambled sgRNA#1 (Synthego) were incubated with Alt-R S.p.HiFi Cas9 Nuclease V3 Cas9 (Integrated DNA Technologies) to form sgRNA–Cas9 ribonuclear protein complexes before electroporating into naive MACS-sorted CD8+ T cells labelled with CTV using a Lonza 4D-Nucleofector system (DS137). Cells were rested overnight before activation in culture for 3 days by plate-bound anti-CD3ε and soluble anti-CD28 (2 µg ml−1 each; BioLegend) and rhIL-2 (5 ng ml−1) in lipid-free medium containing 5 μM U46619 or vehicle control.
RNA-seq analysis
Lung tissues were immediately immersed in the RNAlater Stabilization solution (Thermo Fisher Scientific) for storage at −80 °C. The whole tissue was homogenized in 1 ml RLT Plus lysis buffer (Qiagen) by using a Qiagen TissueLyser II homogenizer and RNA was isolated by using the RNeasy Plus Mini kit (Qiagen) according to the manufacturer’s instructions. Naive CD8+ T cells (CD62L+CD44−CD8+) were purified by total CD8+ T cells enrichment followed by FACS, as described above and activated by plate-bound anti-CD3 and soluble anti-CD28 (5 µg ml−1 each) in lipid-free medium containing 10% Charcoal-Stripped FBS for 5 days in the presence of recombinant rhIL-2 (5 ng ml−1) and U46619 (1 μM). Cells were then pelleted and resuspended in 40 µl RNAlater Stabilization solution for storage at −80 °C. Processing of cell samples was performed using the QIAshredder Kit (Qiagen) and RNA was then extracted using the RNeasy Plus Mini Plus Kit (Qiagen) according to the manufacturer’s protocol. RNA libraries were prepared by Novogene using the mRNA library preparation kit (poly A enrichment) and sequencing was performed by Novogene on NovaSeq PE150. The FastQ files were then subjected to quality control using FastQC and then alignment to the NCBIM37 Mus musculus genome annotation using the STAR workflow. Differential gene expression analysis was performed on all expressed genes (>20 detected reads) using DESeq256, and differentially expressed genes were further analysed and visualized using R. Expression heat maps were generated with the R package heatmap.
Histopathological analysis of tissues
Lungs were fixed in 4% formalin before being embedded in paraffin. Sections were taken from the centre of the lungs and stained with H&E, using routine histology methods. Slides were genotype-blinded and independently scored for pathological features.
Quantification of urinary prostanoid metabolites
Urine samples from mice that received intravenous injections of B16-F10 cells were collected using metabolic cages. Systemic biosynthesis of prostaglandin E2, prostaglandin D2, prostaglandin I2 and TXA2 was evaluated by quantifying their major urinary enzymatic metabolites: PGEM, PGDM, PGIM and TXM, respectively, using liquid chromatography–tandem mass spectrometry as previously described42,57,58. Urine samples (0.2 ml aliquots) were added with internal standards: tetranor PGEM-d6 (final concentration of 10 ng ml−1, Cayman), tetranor PGDM-d6 (final concentration of 10 ng ml−1, Cayman), 2,3-dinor-6-keto-PGF1α-d9 (sodium salt) (final concentration of 10 ng ml−1, Cayman) and 2,3-dinor-TXB2-d9 (final concentration of 10 ng ml−1, Cayman). Samples were incubated at room temperature for 15 min, followed by addition of formic acid (5 μl). After 15 min of incubation, methoxyamine HCl (1 g ml−1, 0.1 ml) (Sigma Aldrich) was added. Following 30 min of incubation at room temperature, urine samples were diluted to 1 ml with water adjusted to pH 3 with HCl and extracted with Number Strata-X 33u polymeric reversed phase (30 mg ml−1, Phenomenex) that had been preconditioned with 1 ml of acetonitrile and 1 ml of water58,59. The Number Strata-X 33u polymeric reversed phase column loaded with samples was washed with 1 ml of water (5% acetonitrile) and then eluted with 1 ml of 5% acetonitrile in ethyl acetate. The eluate was evaporated, and the dried residue was resuspended in 100 μl mobile phase (10% acetonitrile in water), and 30 μl was injected into an ACQUITY UPLC I-Class/Xevo TQS micro-IVD System (Waters) equipped with an electrospray ionization source (ESI Z-Spray), under negative ionization conditions, as previously described58.
Platelet and T cell transwell co-cultures
Platelets were isolated as previously described60. In brief, blood was collected via cardiac puncture using syringes containing 100 μl of acid citrate dextrose (Sigma Aldrich) from B16-F10 bearing mice. Samples were immediately mixed with modified Tyrode’s buffer to prevent premature activation of platelets. Blood was centrifuged at 300g for 5 min and the uppermost platelet-rich plasma (PRP) layer was collected. PRP was subsequently centrifuged at 200g for 8 min to increase purity. Prostacyclin I2 (Sigma Aldrich) was added at a final concentration of 1 μg ml−1 to limit premature platelet activation and the sample was immediately centrifuged for 11 min at 1,000g. The cells were resuspended in modified Tyrode’s buffer. Platelet count and purity were determined by flow cytometry using the following antibodies: Armenian hamster anti-mouse/rat CD61-PE antibody (2C9.G2, BioLegend), rat anti-mouse Ter119-FITC antibody (TER119, eBioscience) and anti-mouse CD45.2-PerCp-Cy5.5 antibody (Ly5.2). Platelet purity was greater than 90%.
Platelets and naive FACS-sorted CD8+ T cells labelled with 2.5 μM CTV were co-cultured with a ratio of 30:1 in 24-well transwell plates (0.4 μm pore size, Corning). Naive CD8+ T cells on the bottom of the transwell were activated by plate-bound anti-CD3ε and soluble anti-CD28 (5 µg ml−1 each) in a lipid-free medium in the presence of rhIL-2 (5 ng ml−1) as described above. After 5 days, supernatants were carefully collected and TXB2 levels were measured using a Thromboxane B2 ELISA Kit (Cayman) according to the manufacturer’s instructions. T cells were collected and processed for flow cytometry as previously described.
Confocal immunofluorescence microscopy
CD8+ T cells from wild-type and Arhgef1-KO OT-1 TCRtg mice were generated in vitro for 5 days as described above. 5 × 105 serum-starved cells were seeded on 13-mm coverslips (VWR) pre-coated with 10 μg ml−1 Ultra-LEAF™ purified anti-mouse CD3ε in a 24-well plate. Cells were stimulated with 10 μM U-46619 or DMSO control for 10 min at 37 °C. After fixation and washing with 15 mM glycine and PBS. Coverslips were blocked with Fc blocking anti-FcR antibody (clone 2.4G2, BD). Cells were then incubated with the rabbit anti-mouse PTEN (138G6, Cell Signalling) antibody, followed by the donkey anti-rabbit IgG-Alexa Fluor 647 (A31573, Thermo-Fischer) antibody and Coralite 594-Phalloidin (Proteintech). Coverslips were mounted onto VWR SuperFrost Microscopy Slides (Appleton Woods MS527) using ProLong Diamond Antifade Mountant with DAPI (Thermo-Fischer P36962). For inhibition experiments, cells were pre-treated with 10 μM TP inhibitor SQ 29548, ROCK inhibitors 30 μM Y-27632 and 10 μM GSK269962A or DMSO for 1 h before seeding.
Images were captured on a Leica TCS SP8 inverted confocal microscope using the Leica Application Suite X (LAS X) software (v1.4.6.28433) with a 63× oil-immersion objective lens, in three channels, F-actin (yellow) and PTEN (red) plus a DAPI nuclear counterstain (blue) to locate the cells. z-stacks were obtained to capture the entirety of cells in each region. Three to five distinct locations were imaged for each replicate of each treatment condition. Images analysed were maximum intensity projections of four consecutive z-slices selected from the centre of a cross-volume image stack. An image analysis pipeline was constructed using Cellprofiler 461. In brief, cell masks were generated from the F-actin image using as a seed a nuclei mask segmented from the DAPI image. These cell masks could be further compartmentalized into membrane and cytoplasmic regions by using the F-actin image. Ratiometric measurements of mean fluorescence intensity of PTEN on the membrane versus intracellular were used to report its re-localization to the membrane.
Single-sample gene set enrichment analysis
Every expressed gene detected in the RNA-seq experiment in Fig. 4a (14,025 genes; Supplementary Table 3) was subject to single-sample Gene Set Enrichment Analysis (ssGSEA) analysis62. ssGSEA was used to test the relative enrichment of each gene set comprising the MsigDB C7 Immunologic signature gene sets in the total expressed gene transcriptional profile of each replicate sample. ssGSEA first calculates for each sample the differential expression of each expressed gene between that sample and the remainder of the samples. The list of expressed genes in each sample is then ranked by their level of differential expression compared to the other samples. The non-random distribution (enrichment) of genes within each C7 gene set for each sample is then calculated, yielding a sample enrichment score for each gene set per sample. This is achieved by analysing the distribution of the genes in each C7 gene set in the list of expressed genes rank ordered by their differential expression for each sample. The sample enrichment score for each differentially enriched gene set (false discovery rate (FDR) < 0.2, |FC| > 1.5) is represented in the heat map provided and the identity of each gene set in the heat map is given in Supplementary Table 4. Each gene within each of the gene sets included in the analysis can be looked up using the following link: https://www.gsea-msigdb.org/gsea/msigdb/human/genesets.jsp?collection=C7.
Cell line authentication
Validated B16-F10 melanoma cells were obtained from ATCC. Plat-E cells were obtained from Cell Biolabs. Validated MC38 colorectal adenocarcinoma cells were obtained from Kerafast. All lines were validated as mycoplasma-free by suppliers and expanded at low passage frequency before cryopreservation.
Statistical testing
Data were analysed using unpaired two-tailed Student’s t tests, Mann–Whitney U-test, ordinary one-way ANOVA or two-way ANOVA test where stated. Most experiments did not require blinding since objective quantitative assays, such as flow cytometry, were used. For tumour experiments littermate controls or age- and sex-matched mice of different genotypes were randomized and the operator blinded to genotype before injection and again before counting of metastatic nodules or assessment of histology images to allow for objective assessment. Experimental sample sizes were chosen using power calculations, preliminary experiments, or were based on previous experience of variability in similar experiments. Samples which had undergone technical failure during processing were excluded from subsequent analysis.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.