Animals
Mice (8–40 weeks old) were housed under controlled light conditions (12 h light/12 h dark) and temperature (22 °C) and fed ad libitum on standard mouse chow. Mice were randomized to treatment group on the basis of body weight. Unless noted otherwise, all mice were male. Mice were group housed except for studies for food intake measurements. Mice used were: B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J, with Cre-dependent tdTomato (Ai14; Jax 007914)45, Vglut2-IRES-cre (Slc17a6tm2(cre)Lowl/J) (Jax 016963)46; Vgat-IRES-cre (Slc32a1tm2(cre)Lowl/J) (Jax 028862)46 and C57BL/6 J (Jax 000664). All mice were housed in a temperature-controlled environment (20–22 °C, 50–60% humidity) with 12 h of light per day at the Center for Comparative Medicine and Surgery (CCMS) at Icahn School of Medicine at Mount Sinai (New York, NY, USA). Animal care and experimental procedures were performed with the approval of the Animal Care and Use Committee of Icahn School of Medicine at Mount Sinai under established guidelines.
General surgical procedures
All surgeries were performed under aseptic conditions. Mice were anaesthetized using 2% isoflurane and the top of the head was shaved then cleaned with 70% ethanol. Ophthalmic ointment was applied to the eyes and subcutaneous injections of buprenorphine (0.05 mg kg−1) were given to each mouse prior to surgery. An incision was made on the midline and small craniotomies were made using a dental drill. Thirty-three gauge syringe needles (Hamilton) were used to unilaterally or bilaterally infuse virus into the brain at a rate of 0.1 µl min−1. The following volumes and coordinates were used: MeA, 0.3–0.5 µl, 1.4 mm posterior, 2.5 mm lateral (2.55 mm if mouse body weight >25 g, 2.6 mm if mouse body weight >30 g), and 5.35 mm ventral from bregma; VMH, 0.3 µl, 1.2 mm posterior, 0.23 mm lateral, and 5.6 mm ventral from bregma; BNST, 0.3 µl, 0.2 mm anterior, 0.85 mm lateral, and 4.3 mm ventral from bregma. Viral expression was confirmed after euthanasia using a fluorescent Zeiss Axio Observer Z.1 microscope to visualize fluorophores and confirm targeting. Mice with misplaced injections or without virus expression were not included in the analysis.
Transneuronal circuit analysis was performed using PRV expressing enhanced green fluorescent protein (GFP) (PRV152). PRV-GFP was injected into the liver of Ai14 mice via a Hamilton syringe (5× 100 nl, 3.96 × 109 pfu ml−1). Seven days after the PRV-GFP injections, mice were euthanized via perfusion and brains dissected and sectioned to visualize PRV-GFP expression.
Viral vectors
We used the following viruses: AAV8-hSyn-hM3D(Gq)-mCherry (gift from B. Roth, Addgene viral prep #50474-AAV8; RRID:Addgene_50474); AAV8.2-synapsin-mCherry (Virovek); AAV8.2-hEF1a-synaptophysin-mCherry (Massachusetts General Hospital Gene Delivery Technology Core, AAV-RN8, RRID:SCR_012544); AAV9-hSyn-FLEX-mGFP-2A-Synaptophysin-mRuby (Addgene viral prep #71760); AAV/retro-RFP (gift from K. Deisseroth, Addgene viral prep #114472-AAVrg, RRID:Addgene_114472); AAV/retro-GFP (gift from B. Roth, Addgene viral prep #50465-AAVrg, RRID:Addgene_50465); AAV2/retro-CAG-Cre-WPRE (Boston Children’s Hospital Viral Core); AAV8-hSyn-DIO-hM3D(Gq)-mCherry (gift from B. Roth, Addgene viral prep #44361-AAV8; RRID:Addgene_44361); AAV8-hSyn-DIO-mCherry (gift from B. Roth, Addgene viral prep #50459-AAV8; RRID:Addgene_50459); AAV8-EF1a-mCherry-flex-dtA (Canadian Neurophotonics Platform Viral Vector Core Facility, RRID:SCR_016477)47; AAV1-hSyn-Cre (gift from J. M. Wilson, Addgene viral prep #105553-AAV1, RRID:Addgene_105553); AAV9-CaMKIIa-hM3D(Gq)-mCherry (Addgene prep #50476); pAAV-hDlx-GqDREADD-dTomato-Fishell-4 (Addgene prep #83897); AAV9-syn-jGCaMP8s-WPRE (Addgene viral prep #162374); AAV9-hSynapsin1-axon-jGCaMP8s-P2A-mRuby3 (Addgene viral prep #172921); AAV9-hSyn-hChR2(H134R)-EYFP (Addgene viral prep #26973); AAV9-hSyn1-EYFP (Addgene viral prep #117382); PRV152 (gift from L. Enquist)48.
Stereotaxic injection and fibre optic implantation
Mice were anaesthetized with 2% isoflurane and placed in a stereotaxic head frame (Kopf Instruments). Ophthalmic ointment was applied to the eyes and subcutaneous injections of meloxicam (5 mg kg−1) and Enrofloxacin (5 mg kg−1) were given to each mouse prior to surgery. The scalp was shaved and scrubbed with iodine and alcohol and an incision made on the midline. A craniotomy was made using a dental drill (0.5 mm) above the MeA, VMH or BNST.
For photometry experiments, a craniotomy was made above the MeA at the following coordinates AP: −1.4 mm, ML: +2.5 mm, DV: 5.35 mm from bregma. A 0.3 µl volume of AAV9-syn-jGCaMP8s-WPRE was injected at a rate of 100 nl min−1 using a 10 µl Hamilton syringe controlled by a micro-injector. The needle remained in the injection site for 2 min following completion of delivery before being raised 0.1 mm for a further 2 min before being completely retracted. A fibreoptic cannula (MFC_400/430-0.66_6mm_MF1.25_FLT) (Doric) was implanted 0.2 mm dorsal to viral injection during the same surgery and was secured to the skull using dental cement (Pearson Dental) and three screws (Plastics One).
For axon-specific calcium imaging, a craniotomy was made above the MeA at the following coordinates AP: −1.4 mm, ML: +2.5 mm, DV: 5.35 mm. 0.3 µl of AAV9-hSynapsin1-axon-jGCaMP8s-P2A-mRuby3 was injected at a rate of 100 nl min−1 using a 10 µl Hamilton syringe controlled by a micro-injector. The needle remained in the injection site for 2 min following completion of delivery before being raised 0.1 mm for a further 2 min before being completely retracted. A second craniotomy was made using a dental drill (0.5 mm) at the following coordinates: VMH 1.2 mm posterior, 0.23 mm lateral, and 5.6 mm ventral from bregma; BNST 0.2 mm anterior, 0.85 mm lateral, and 4.3 mm ventral from bregma. A fibreoptic cannula (MFC_400/430-0.66_6mm_MF1.25_FLT) (Doric) was implanted 0.2 mm dorsal to region of interest during the same surgery and was secured to the skull using dental cement (Pearson Dental) and three screws (Plastics One).
For optogenetic stimulation of MeA cell bodies, a craniotomy was made using a dental drill (0.5 mm) at the following coordinates AP: −1.4 mm, ML: +2.5 mm, DV: 5.35 mm. 0.3 µl of pAAV9-Syn-ChR2(H134R) was injected at a rate of 100 nl min−1 using a 10 µl Hamilton syringe controlled by a micro-injector. The needle remained in the injection site for 2 min following completion of delivery before being raised 0.1 mm for a further 2 min before being completely retracted. A 1.25 µm, NA 0.66 fibreoptic (MFC_400/430-0.66_6mm_MF1.25_FLT) (Doric) was implanted unilaterally 0.2 mm dorsal to viral injection during the same surgery.
For stimulation of MeA projections, optical fibres were unilaterally implanted over MeA terminal regions at the following coordinates VMH 1.2 mm posterior, 0.23 mm lateral, and 5.6 mm ventral from bregma; BNST 0.2 mm anterior, 0.85 mm lateral, and 4.3 mm ventral from bregma. Fibreoptics were secured to the skull using dental cement (Pearson Dental) and three screws (Plastics One). Mice were allowed at least 6 weeks for recovery and to facilitate sufficient viral expression prior to any experimental procedures.
Fibre photometry
Mice were tethered to a patch cable (Doric Lenses, MFP_400/430/1100-0.57_3m_FCM-M1.25). Calcium signals were collected using the Doric Fluorescence MiniCube and fibre photometry console at a sampling frequency of 12 kHz using Synapse software (v.98, Tucker-Davis Technologies). GCaMP calcium signal (465 nm) and UV isosbestic signal (405 nm) were collected through the same fibre and equalized to record an equivalent signal:noise ratio. MATLAB (v.2022b, Mathworks) scripts provided by Tucker-Davis Technologies were used to down-sample and normalize the fluorescence signal. For projection specific calcium imaging, photobleaching was corrected using double exponential fit. The 405 isobestic fluorescence signal was filtered using a polyfit regression giving a fitted control (F405c). ΔF/F was calculated by subtracting F405c from the GCaMP fluorescence signal (F405) and then dividing by F405c(F465 – F405c)/F405. A z-score conversion was used to calculate the deviation of the resulting ΔF/F from the averaged signal of the entire recording session.
Restraint stress
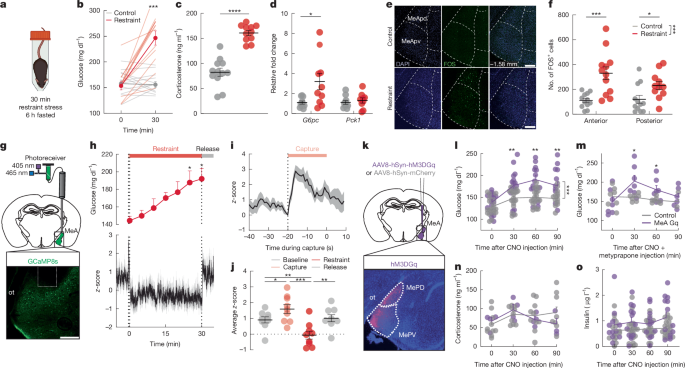
Mice were tethered to the patch cable, and a 5 min baseline recording was collected. Mice were then restrained in plastic 50-ml conical tubes (Fisher Scientific) with holes that were modified to account for the patch cable tethering. Calcium transients were recorded for 5 or 30 min while the mouse remained secured. After the restraint, the mouse was released and recording continued for an additional 15 min. When noted, blood glucose was measured every minute for the duration of a 5 min restraint stress or every 5 min for the duration of a 30 min restraint stress to assess the time course of stress-induced hyperglycaemia.
Territorialized cage stress
Mice were tethered to a patch cord, placed in a clean novel cage with bedding, and recording started. A 5 min baseline was collected in the ‘clean’ novel cage before the mouse was manually picked up by the base of the tail and placed in a novel territorialized cage that was previously occupied by 5 males for 1 week. The mice remained in this cage while calcium transients were recorded for 5 or 30 min before the mouse was placed back in the previous clean cage for 5 min. To examine the effects of repeated stress, a cycle of 2 min of territorialized cage exposure, followed by 3 min of clean cage exposure was repeated for a total of 5 novel territorialized cage exposures. To examine the effects of intermediate stress, mice were exposed to a novel territorialized cage for 5 min at intervals of 0 h, 6 h, 12 h and 24 h from the initial territorialized cage exposure. When noted, blood glucose was measured every minute for the duration of a 5 min territorialized cage stress or every 5 min for the duration of a 30 min territorialized cage stress to assess the time course of stress-induced hyperglycaemia.
Footshock stress
To examine the effect of an acute physical stressor, mice were exposed to a mild footshock. Mice were tethered to a patch cord and placed into a fear conditioning chamber. A 5 min baseline was collected before a 1 s, 1 mA footshock was delivered. The calcium transients were recorded during the shock and post shock for at least 2 min. We scored the calcium response at the time of the shock.
Robobug stress
To examine the effect of an acute visual stressor, mice were tethered to a patch cord and placed into an open field arena. After 5 min, a remote-controlled spider (robobug) was introduced into the open field and began to pursue the mouse at random intervals. The calcium transients were recorded during the approach and post-approach for at least 1 min. We scored the calcium response at the time of the approach.
Continuous glucose monitoring
Continuous glucose monitoring was performed in a subset of mice during calcium imaging and stress exposure. In brief, under isoflurane anaesthesia, hair was removed from the flank of the mouse and a 2 mm incision made in the skin. The cannula of the continuous glucose monitor (Abbott Freestyle Libre 2) was inserted into the incision and the adhesive tape on the monitor firmly applied to the skin. Discontinuous sutures around the sensor attached the adhesive tape to the skin. The sensor was activated, and mice were allowed to recover for 24 h before fibre photometry and stress exposure.
Optogenetics
Photostimulation was performed with 10 ms pulses given at 20 Hz for 2 s, repeated every 2 s. The output beam from a 473 nm diode laser (Thorlabs) was controlled by a microcontroller (Uno, Arduino, IDE 2.3.1) running a pulse generation script. The laser was coupled to a 200 µm, NA 0.66 multimode optical fibre (MFC_400/430-0.66_6mm_MF1.25_FLT) (Doric) using a 1.25 mm mating sleeve (Thorlabs) that allowed delivery of the light into the brain by coupling to the implanted fibre optic. Laser power was measured using an optic power meter (Thorlabs) and set at 5 mW.
Metabolic studies
In each experiment, mice were tethered to a patch cable, placed in an open field arena, and habituated for at least 1 h to account for the handling stress. For basal glucose measurements during optogenetic modulation, mice were fasted for 6 h and tail vein samples for blood glucose were taken at 0, 5, 15 and 30 min after the initiation of stimulation. To measure tolerance to a glucose challenge, mice were fasted for 6 h and tail vein samples for blood glucose were taken at 0, 15, 30, 45 and 60 min after injection of glucose (2 mg kg−1 body weight) and the initiation of stimulation. To measure gluconeogenic capacity, mice were fasted for 4 h and tail vein samples for blood glucose were taken at 0, 15, 30, 45 and 60 min after intraperitoneal injection of pyruvate (2 g kg−1 body weight, Sigma P5280). Data are shown as the change in blood glucose from baseline (immediately before stimulation).
Stress studies
In each experiment, mice were tethered to a patch cable, placed in an open field arena, and habituated for at least 1 h to account for the handling stress. Mice were exposed to both restraint and territorialized cage stress as described above. For stressed glucose measurements during optogenetic modulation, mice were fasted for 6 h, and tail vein samples for blood glucose were taken at 0, 5, 15, 30 min. For 5 min stressors, the mice were optogenetically stimulated for 5 min, beginning at the initiation of stress. For 30 min stressors, the mice were optogenetically stimulated for 15 min, beginning at the initiation of stress.
In vivo chemogenetic behavioural testing
Mice were handled for 5–10 days before experiments. Following stereotaxic surgeries, mice were allowed to recover for 3–6 weeks before the start of testing. Where applicable, CNO (Sigma, NIH) was dissolved in 10% DMSO in saline and delivered at a dose of 3 mg kg−1, intraperitoneal injection. Investigators were blinded to treatment groups.
Restraint stress
Mice were fasted for 6 h and then either briefly handled and returned to home cage (controls) or restrained in a 50-ml falcon tube with a hole cut for air at the conical end for 5 or 30 min. For 5 min restraint experiments, blood glucose was measured before the initiation of stress, after the 5 min restraint, and 30 min after the initiation of the stress. For 30 min restrain experiments, blood glucose was measured before and after the 30 min restraint period. To measure the hormonal or gene expression responses to stress, mice were rapidly anaesthetized with 3% isoflurane at the end of the restraint period and blood or tissue collected. To measure FOS in the MeA after restraint stress, mice were anaesthetized with 3% isoflurane and perfused transcardially with 0.1 M phosphate buffered saline (PBS) followed by 10% formalin, and the brain was removed 2 h after the start of the restraint. For DREADD modulation studies, CNO was administered 30 min before restraint.
Repeated restraint stress
To determine the effects of chronic repeated restraint stress on blood glucose, blood glucose was measured before and after a 30 min restraint stress at 12 h intervals for a total of 9 restraint sessions. To measure FOS in the MeA after chronic restraint stress, mice were anaesthetized with 3% isoflurane and perfused transcardially with PBS followed by 10% formalin, and the brain was removed 2 h after the start of the restraint.
Territorialized cage stress
Mice were placed in a dirty cage previously occupied by five male mice for one week. Blood glucose or food intake was measured before and after the 5 or 30 min period of territorialized cage exposure. For blood glucose measurement, mice were fasted for 6 h. To measure food intake, mice were food-deprived overnight before being placed in the territorialized cage. For DREADD modulation studies, CNO was administered immediately before the test.
Food intake studies
Mice were food-deprived overnight or food-deprived for 6 h in the light phase or allowed to eat ad libitum. Food, either in the form of standard rodent chow or palatable food (peanut butter) was then provided in excess, and consumption of food was measured every hour. For DREADD modulation studies, CNO was administered immediately before food was provided.
Metabolic studies
For baseline glucose measurements after DREADD modulation, mice were fasted for 6 h, and tail vein samples for blood glucose were taken at 0, 30, 60 and 90 min after intraperitoneal injection of CNO. To measure tolerance to a glucose challenge, mice were fasted for 6 h and tail vein samples for blood glucose were taken at 0, 10, 20, 30, 45, 60, 90 and 120 min after injection of glucose (2 mg kg−1 body weight). When noted, additional blood was collected at 0, 10, 30, 60 and 90 min after glucose injection to measure plasma insulin and glucagon. To measure insulin sensitivity and tolerance to an insulin challenge, mice were fasted for 4 h and tail vein samples for blood glucose were taken at 0, 30, 60, 90 and 105 min after injection of insulin (0.4–0.6 U kg−1 body weight, Humulin R HI-210). To measure gluconeogenic capacity, mice were fasted for 4 h and tail vein samples for blood glucose were taken at 0, 30, 45, 60, 90 and 105 min after intraperitoneal injection of pyruvate (2 g kg−1 body weight, Sigma P5280). For all metabolic challenges, CNO was injected 30 min before the challenge (timepoint 0).
Open field activity
Locomotion and anxiety-like behaviour were measured in either a clear plexiglass 40 × 40 × 30 cm open field arena using Fusion Software (v5.0) (Omnitech Electronics) or a white acrylic 18 × 18 × 18 in arena using Ethovision XT (version xt13, Noldus Information Technology) to quantify behaviour. Distance travelled and time spent in the center of the open field arena were measured. Open field testing lasted 10 min. For DREADD modulation studies, CNO was administered 60 min before the start of the test.
Elevated plus maze and light-dark box
The light-dark box test was performed in a 40 × 40 × 30 cm arena with a black box placed on half of the arena to shield it from light. The mice were placed in the light portion and tracked using Fusion Software. For the elevated plus maze, the mice were placed on an open arm of an elevated four-arm maze in which two arms are open and two are enclosed. They were tracked using Ethovision software and total time spent in the open arm was measured. Elevated plus maze and light-dark box testing lasted 10 min.For DREADD modulation studies, CNO was administered 60 min before the start of the test.
High-fat diet
Mice with MeAVMH expression of DTA or mCherry were fed a high-fat diet (Research Diets, D12492; 60% fat). Food intake, body weight and blood glucose were measured every 3–7 days for 20 days.
Metyrapone studies
To pharmacologically block corticosterone production, mice were injected with metyrapone (50 mg kg−1, subcutaneous injection, dissolved in 5% Tween 80, Tocris Bioscience) or vehicle 60 min before the initiation of restraint stress, basal glucose measurements, or glucose administration during GTT. For DREADD modulation studies, CNO was administered 30 min before the start of the test.
Neuronal tracing
To trace MeA projection circuits, an injection of AAV8.2-hEF1a-synaptophysin-mCherry was unilaterally injected into the MeA. After 4–6 weeks to allow viral expression, brains were collected and prepared for imaging as described in the Immunohistochemistry section. Sections from the whole brain were imaged to allow unbiased selection of MeA projection targets. After mCherry-labelled regions were identified, fluorescence intensity was quantified.
To determine axonal projection overlap in the MeA, mice were injected with AAVretro-GFP into the VMH and AAVretro-RFP into the BNST to differentially label MeA neurons that project to the VMH and MeA neurons that project to the BNST. Expression of GFP and of RFP in the MeA was quantified to determine axonal projection overlap.
To determine MeA projection circuit specificity, mice were injected with a AAVretro-GFP into the VMH and AAVretro-RFP into the BNST to differentially label MeA neurons that project to the VMH from MeA neurons that project to the BNST. These mice were exposed to a 30 min restraint stress. To measure FOS in the MeA after restraint stress, mice were anaesthetized with 3% isoflurane and perfused transcardially with PBS followed by 10% formalin, and the brain was removed 2 h after the start of the restraint. Co-expression of the projection specific GFP or RFP and of FOS in the MeA was quantified to determine the response to stress in the MeA→VMH projection and in the MeA→BNST projection.
For polysynaptic circuit tracing of MeAVMH to peripheral organs, Ai14 mice were injected with AAV1-Cre into the MeA. After 4-weeks to allow viral expression, mice were injected with PRV152-GFP into the liver. Seven days after the injection of the PRV-GFP, these mice were perfused, and the brains were collected. Co-expression of monosynpatic anterograde RFP expression from the MeA and of polysynaptic retrograde GFP expression from the liver was quantified in the VMH to determine the overlap of neurons that project from the MeA to the VMH and have a polysynaptic connection to the liver.
Vglut2-cre or Vgat-cre mice were injected with a AAV9-hSyn-FLE-mGFP-2A-Synaptophysin-mRuby to determine cell-type specificity of MeA neurons that project to its downstream regions. Expression of mRuby in Vglut2-cre mice was used to determine the proportion of glutamateric neurons that project from the MeA to the VMH, BNST, lateral hypothalamus and medial preoptic area. Expression of mRuby in Vgat-cre mice was used to determine the proportion of GABAergic neurons that project from the MeA to the VMH, BNST, lateral hypothalamus and medial preoptic area.
Cell-type-specific chemogenetic activation
Mice were injected with a AAV9-CaMKIIa-hM3D(Gq)-mCherry, a AAV9-hDlx-GqDREADD-dTomato-Fishell-4, or a AAV9-hSyn-mCherry into the MeA. Mice were allowed 6 weeks for viral expression. For baseline glucose measurements after DREADD modulation, mice were fasted for 6 h, and tail vein samples for blood glucose were taken at 0, 30, 60, 90 min after intraperitoneal injection of CNO (3 mg kg−1). The response to DREADD activation driven by the CAMKIIa promoter or driven by the Dlx promoter was compared to mCherry-expressing control mice to determine the contribution of excitatory or GABAergic neurons to changes in blood glucose.
Tissue processing
Blood glucose was determined using a Contour or Contour Next EZ glucometer (Bayer).
Blood for plasma hormones was collected in an EDTA-coated tube (Sarstedt Microvette CB 300 K2E 16.444.100), spun for 10 min at 2,000 rpm, 4 °C, and plasma was separated and stored at −80 °C until assay. Plasma levels of insulin (Mercodia 10-1247-01), glucagon (Crystal Chem 81518), corticosterone (Crystal Chem 80556), adrenaline and noradrenaline (Abnova KA1877) were determined by ELISA. Liver glycogen was determined by colorimetric assay (Abcam ab169558). Circulating glycerol and triglyceride levels were measured by enzymatic assay (Sigma-Aldrich TR0100). SpectraMax i3x, (Molecular Devices) with SoftMax Pro (v.7.1.2) were used for ELISA detection.
To examine protein levels or gene expression, liver was collected immediately after 30 min restraint or territorialized cage stress. For DREADD modulation studies, CNO was administered 60 min before the collection of liver tissue. MeAVMH hM3DGq-expressing and corresponding control mice were unstressed. MeAVMH hM4DGi and corresponding control mice were 4 h fasted and exposed to a 30 min restraint stress with liver tissue collected immediately after the restraint stress. Liver tissue was flash frozen in liquid nitrogen for gene expression analysis, glycogen and protein analyses, aliquoted and stored at −80 °C until processing.
Western blot
Liver tissue (~20 mg) was lysed in 600 µl buffer (20 mM Tris, pH 7.4, 150 mM NaCl, 2% Nonidet P-40, 1 mM EDTA, pH 8.0, 10% glycerol, 0.5% sodium deoxycholate, 0.2% semi-dehydroascorbate) supplemented with halt protease and phosphatase inhibitor (Cell Signaling). Livers were first homogenized using Beadbug (Benchmark) and lysates were further sonicated in ice-cold water for 5 min. After centrifuging at 14,000 rpm at 4 °C for 10 min, the protein supernatants were transferred to a new tube and protein concentration was determined by BCA protein assay kit (Pierce). Thirty micrograms of protein was mixed with 6× SDS sample buffer (BP-11R, Boston BioProducts) and boiled for 5 min before loading on SDS–PAGE. The Bio-Rad wet transfer system was used to transfer proteins for western blotting. PVDF membranes were blocked with 5% dry milk in TBST (TBS + 0.05% Tween 20) and further incubated with primary antibodies (dilute in TBST with 3% BSA + 0.05% NaN3) at 4 °C overnight. PCK1 (ab70358), PGC1a (ab54481), FoxO1 (ab39670), GAPDH (ab9485) antibodies were from Abcam. G6PC (NBP1-80533) was from Novus Biologicals. All primary antibodies were used at 1:1,000 dilution. The membrane was washed 4 times in TBST with shaking for 10 min prior to incubation with secondary antibodies (HRP Goat anti-rabbit IgG (Thermofisher, 31460, 1: 10,000) and HRP Goat anti-mouse IgG (Thermofisher, 31430,1: 10,000 in TBST)) for an additional 2 h at room temperature. Immune complexes were washed 4 times in TBST with shaking for 10 min at room temperature. Membranes were further reacted with Pierce ECL western blotting substrate and imaged with iBright CL1500 (Thermofisher). Western blots were quantified by Image J (v.1.53k).
Quantitative PCR
Total RNA was extracted from tissue by homogenization in Trizol (Invitrogen) followed by chloroform (Sigma) extraction and isolation using the RNeasy Plus Mini (Qiagen) kit according to manufacturer’s instructions. Complimentary DNA was prepared by reverse transcription of 500 ng total RNA using qScript cDNA SuperMix (Quantabio). The resulting cDNAs were diluted 1:10 then amplified by real-time PCR using the SYBR green system (Applied Biosystems) according to the manufacturer’s protocols. Data were analysed with Quantstudio Design and Analysis software (v.1.5.2). All mRNA expression data were normalized to Rpl23 expression in the corresponding sample. Fold change in mRNA expression was calculated using the delta-delta Ct method49. The follow primers were used: Pck1 forward GCGAGTCTGTCAGTTCAATACC, reverse GGATGTCGGAAGAGGACTTTG; G6Pc forward GGAGGCTGGCATTGTAGATG, reverse TCTACCTTGCTGCTCACTTTC; Rpl23 forward ACTTCCTTTCTGACCCTTTCC, reverse TTAGCTCCTGTGTTGTCTGC; Pgc1a forward TGAGGACCGCTAGCAAGTTT, reverse TGTAGCGACCAATCGGAAAT; FoxO1 forward GCGTGCCCTACTTCAAGGATAA, reverse TCCAGTTCCTTCATTCTGCACT; Glut2 forward GTTGGAAGAGGAAGTCAGGGCA, reverse ATCACGGAGACCTTCTGCTCAG; Irs2 forward CCAGTAAACGGAGGTGGCTACA, reverse CCATAGACAGCTTGGAGCCACA; Igfbp1 forward GCCCAACAGAAAGCAGGAGATG, reverse GTAGACACACCAGCAGAGTCCA; Adra1a forward CTAAGGCCATTCTACTTGGGGT, reverse CGAGTGCAGATGCCGATGA; Adra1b forward GTCGGAATGTTCATCTTATGTTGG, reverse CAGCCAGAACACTACCTTGA; Adra2a forward CCTGCTTTGACATTTCCTGAC, reverse TCATTTCCTTCTGCCTTGGTC, Adra2b forward AGACCTCATCCTCAGACACC, reverse TCCAAGCTACCCTTCCTGAA; Adra2c forward GCTGACTTCCTATGACCTGAA, reverse TTGGCTGTCATTGTATTTGGC, Adrb1 forward CTCATCGTGGTGGGTAACGTG, reverse ACACACAGCACATCTACCGAA; Adrb2 forward TCGCTATGTTGCTATCACATCG, reverse GCCAGATACAATCCATACCATCA; Adrb3 forward GGCCCTCTCTAGTTCCCAG, reverse TAGCCATCAAACCTGTTGAGC.
Stable flux isotope analysis
Optogenetic mice that either expressed AAV9-hSyn-hChR2(H134R)-EYFP or AAV9-hSyn1-EYFP were tethered to a patch cable and habituated to an open field arena for at least 1 h. The mice were pre-stimulated for 15 min, administered with a 2 g kg−1 intraperitoneal injection of labelled [2,3-13C]pyruvate (Sigma 490717), and stimulated for 15 min. Immediately following the period of stimulation, the liver was collected and flash frozen.
Liver tissue was collected to determine the flux through the gluconeogenic metabolic pathway by measuring how the stable isotope is incorporated into metabolites. Liver cells were isolated and incorporated into a media, collected to a screw vial, and snap frozen in liquid nitrogen. Following thawing on ice, 200 μl of the media was added to 800 μl of methanol. After a brief vortex, the media was centrifuged at 13,100g for 10 min at 4 °C. The supernatant was transferred to a glass vial and dried under gentle nitrogen flow. The samples were then subjected to a 2-step derivatization with 50 μl of methoxyamine hydrochloride (MOA, 15 mg ml−1 in pyridine) at 30 °C for 90 min, and then incubated with 50 μl of N,O-bis(trimethylsilyl)trifluoroacetamide (BSTFA, containing 1% TMCS) at 70 °C for 60 min. The samples were analysed by gas chromatography–mass spectrometry (GC (8890)-MS (5977B), Agilent). The data was first processed with MassHunter software (v.10.2, Agilent). The mass-to-charge ratio used for the metabolites are 245 for fumarate, 465 for citrate, 335 for malate, 334 for aspartate, 445 for glycerol-3-phosphate, 160 for the glucose fragment of carbon position 1 and 2, and 319 for the glucose fragment of carbon 3 to carbon 6. Isotope enrichment calculation (background correction) was performed as described50.
Isotope enrichment calculations were performed, including correction of measured mass isotopomer distributions for contributions from natural isotope abundances51. Glucose mass isotopomers in glucose, and glucose metabolites (glycolytic and TCA cycle intermediates) are reported as molar fractions of M0, M1, M2, etc., according to the number of labelled carbons in the molecule52. The sum of all isotopomers of the molecules, mi for 1 to n (n = 3, 5 or 6 for pyruvate, citrate or glucose, respectively), equal to 1 or 100%. The labelled isotopomer fractions consist of a distribution not affected by the dilution with unlabelled compounds, reported as mi/ Smi, as a percentage. The enrichment sigma Mn, (SMn) is the weighted sum of the labelled species (SMn = 1M1 + 2M2 + 3M3 + 4M4 + 5M5 + 6M6) = average number of 13C carbons per molecule = 6 for U13C glucose (SMn with only M6 nonzero, and equal to 1) as previously reported52,53.
Immunohistochemistry
Brains
Mice were anaesthetized with 3% isoflurane and perfused transcardially with PBS followed by 10% formalin. The brains were post-fixed in 10% formalin at 4 °C overnight, then washed in PBS at 4 °C overnight. 50 μm coronal slices were cut by vibratome (Leica VT1000). For FOS staining, slices were incubated in blocking solution overnight at 4 °C (3% normal donkey serum (NDS, Sigma) in 0.01% Triton X-100 in 0.01 M PBS (PBT)) and then in primary antibody in blocking solution at 4 °C. The following primary antibodies, concentrations, and incubation periods were used: Cell Signaling rabbit monoclonal anti-FOS (2250), 1:500 for 72 h; abcam chicken polyclonal anti-mCherry (ab205402), 1:1,000 overnight or with Abcam chicken polyclonal to tyrosine hydroxylase (ab76442), 1:500 for 72 h. The slices were then washed in 0.01 M PBS (3× 1 h), incubated in secondary antibody in blocking solution for 2 h at room temperature, and washed in PBS (2× 1 h), with a final wash overnight at 4 °C. The following secondary antibodies and concentrations were used: Jackson Alexa Fluor 647 AffiniPure donkey anti-rabbit (711-605-152), 1:250; Jackson Alexa Fluor 594 AffiniPure donkey anti-chicken (703-585-155), 1:2,000.
For RFP or mCherry staining to enhance endogenous fluorescence of AAV/retro-RFP, AAV8-hSyn-DIO-hM3D(Gq)-mCherry and AAV8.2-hEF1a-synaptophysin-mCherry, slices were washed in 0.01 M PBS (3× 10 min), incubated in blocking solution (3% NDS in 0.01% PBT) for 1 h at room temperature, incubated in primary antibody (Rockland rabbit polyclonal anti-RFP (600-401-379), 1:1,000) overnight at 4 °C, washed in 0.01% PBT (3× 10 min), incubated in secondary antibody (Invitrogen donkey anti-rabbit Alexa Fluor 594 (A-21207), 1:500), and washed in PBS (3× 10 min). For GFP staining to enhance endogenous fluorescence of AAV/retro-GFP or PRV-GFP, slices were washed in 0.01 M PBS (3× 10 min), incubated in blocking solution (3% NDS in 0.01% PBT) for 1 h at room temperature, incubated in primary antibody (Abcam goat polyclonal anti-GFP (ab5450), 1:1,000) overnight at 4 °C, washed in 0.01% PBT (3× 10 min), incubated in secondary antibody (Invitrogen donkey anti-goat Alexa Fluor 488 A-11055, 1:500) and washed in PBS (3 ×10 min).
Locus ceruleus
Brainstems were sectioned from MeA→VMHhM3DGq or mCherry control mice euthanized 2 h after CNO administration. The sections were stained as described above for the expression of tyrosine hydroxylase and for the expression of FOS.
Coeliac ganglia
The coeliac ganglia were dissected from MeA→VMHhM3DGq or mCherry control mice euthanized 2 h after CNO administration. The tissue was post-fixed in 10% formalin at 4 °C overnight, cryo-protected in 30% sucrose (Sigma-Aldrich, 50389) in PBS, embedded in O.C.T. Compound (Thermofisher Scientific, Watham, MA; 23-730-572), frozen at −80 °C, and sectioned at 10 µm thickness. Slides were washed in 0.03% PBT (3× 5 min), incubated in blocking solution overnight at 4 °C (2% normal donkey serum, 3% bovine serum albumin in 0.03% PBT), incubated in primary antibodies for 48 h at 4 °C (Cell Signaling anti-FOS, 1:100; abcam chicken polyclonal to tyrosine hydroxylase (ab76442), 1:500), washed in 0.03% PBT (3× 5 min), incubated in secondary antibodies for 2 h at room temperature (Jackson AF-647 donkey anti-rabbit, 1:250; Jackson AF-594 donkey anti-chicken, 1:500), and washed in 0.03% PBT (3× 5 min). After staining, tissue sections were mounted with DAPI counterstain (Fluoromount).
Verification of viral expression and fibre placement
Mice were transcardially perfused with PBS followed by 10% formalin. Brains were removed and post-fixed overnight in formalin at 4 °C and then washed overnight in PBS at 4 °C. Coronal brain sections were cut (50-μm sections) on a vibratome. The brain sections were stained according to the above protocol. Confocal images were taken using Zeiss LSM 780 confocal microscope using Zen software (Zen 2012 SP5) to verify fibre placements and viral expression.
Image quantification
All confocal images were taken at 20× and tiled. All image analyses were performed using Image J (v.1.53k) or Fiji (v.2.14.0) with JaCOP plugin. Investigators were blinded to treatment groups for FOS analyses.
Synaptophysin-mCherry
4 weeks after stereotactic surgery, mice were perfused, and brains were sliced and stained to enhance mCherry staining. Confocal images were then taken using a Zeiss LSM 780 confocal microscope. Regions of interest (ROI) were drawn on the basis of DAPI staining and the Allen mouse brain atlas54 (https://mouse.brain-map.org/static/atlas) and Franklin and Paxinos mouse brain atlas55 (https://labs.gaidi.ca/mouse-brain-atlas/). The same selection was used for each brain region to normalize for area analysed and fluorescence intensity was measured within the ROI. Values are reported as median pixel intensity ± standard error of the median.
FOS in the brain
z-stack confocal images were taken using an upright LSM 900 (restraint versus control). To measure FOS expression after stress, an ROI was drawn around the MeA complex, including the dorsal, ventral, and basomedial subregions. Images were made binary and cell quantification was performed using the ‘analyze particle’ function. The JaCOP plugin56 was used to measure total expression of FOS after restraint stress or control, overlap of FOS with AAV/retro-RFP (BNST-projecting neurons) and AAV/retro-GFP (VMH-projecting neurons) and overlap of FOS with tyrosine hydroxylase in the locus ceruleus.
FOS in the locus ceruleus and in the coeliac ganglia
z-stack confocal images were taken using a Zeiss LSM 900 using Zen software (Zen 2012 SP5). Tyrosine hydroxylase (TH) expression was used as a mask to select an ROI of only neurons. Then overlap of FOS and DAPI was measured using the JaCOP plugin. Data is reported as number of FOS+ DAPI particles.
Spatial transcriptomics
Spatial transcriptomics was performed using the Xenium assay (10X Genomics). Mice were injected with a AAVretro-mCherry into the VMH. After 4 weeks to allow viral expression, the mice were perfused, and brains were collected. Brains were post-fixed overnight and then paraffin-embedded. In brief, formalin-fixed, paraffin-embedded blocks with coronal embedded mouse brains were sectioned at 5 μm (Leica Histocore biocut), placing 3 brain sections per Xenium slide; serial sections were cut onto on standard charged slides and used for Immunofluorescence Control Slides. Xenium slides were baked for 3 h at 42 °C for section adherence and stored in desiccator until use (no more than 4 weeks). Xenium Slides were deparaffinized and decrosslinked, then processed according to the manufacturer’s V1 protocol using the Mouse Brain panel modified with 100 custom gene targets (Supplementary Data Table 2, S1), hybridized for 20 h, and run on the Xenium instrument.
Post-Xenium slides were stored in 1,000 µl PBS-T (0.05%) for one day at 4 °C. In parallel, immunofluorescence control slides were deparaffinized and decrosslinked using the same conditions and reagents as previously described for Xenium slides. PBS-T was removed, and slides were washed with 500 µl of PBS-T for a total of three washes, 1 min each. Blocking Buffer was prepared as a solution consisting of 1× PBS pH 7.4, 0.1% of Tween- 20, 10% heat-inactivated FBS, and 10 mg ml−1 dextran sulfate and slides were blocked for 1 h at room temperature. Slides were incubated with Rabbit anti-mCherry primary (Rockland 600-401-379 s, 1:500) in blocking buffer overnight at 4 °C. Slides were washed 3× 10 min in PBS-T and then incubated with Donkey anti-rabbit AlexFluor 694 (Jackson AF-647 donkey anti-rabbit) secondary antibody for 2 h at room temperature (1:250). Slides were washed 3 times for 10 min each in PBS-T and mounted in Fluoromount-G (ThermoFisher Scientific 00-4958-02) followed by imaging with a EVOS S1000 imager with EVOS S1000 Spatial Imaging Software (v.1.0, ThermoFisher Scientific).
Sample processing and cell classification using Xenium spatial data
The starting gene by cell raw count matrices and cell segmentation polygons for the 12 spatial transcriptomic samples were generated by Xenium Onboard Analysis. This data was further analysed in R (v.4.4.2), using the Giotto Suite (v.4.2.1)57. Cells with fewer than ten unique genes present were removed. In each sample, the cells corresponding to the two MeA regions were manually selected using the plotInteractivePolygons tool in Giotto. The 24 ROIs contained a total of 21,607 cells. Gene expression normalization was performed using the default Giotto parameters. Cell-type annotation was achieved by Leiden clustering (resolution = 0.25) followed by cell-type-specific marker enrichment analysis (Supplementary Data Table 2, S3). As a result, the initial 15 clusters were merged into 7 cell types with 8 cell clusters annotated as neurons. One cluster that had more than one cell-type signature enriched was annotated as “Other”. The neuronal subset was further subclustered (resolution = 0.5), and a subset of 141 cells (3 smallest clusters) with mixed cell-type signature were removed and relabelled as ‘Other’, leading to a total of 12,512 cells being classified as neurons, distributed across 20 neuronal clusters. Cluster marker genes were identified using scran58 method in Giotto (false discovery rate (FDR) ≤ 0.0001 and log2(fold change) ≥ 0.378). Neuronal subtypes were annotated on the basis of the expression levels of known glutamatergic (Vglut1 and Vglut2) and GABAergic (Vgat) neuronal markers.
mCherry signal analysis and DEGs from Xenium spatial data
Post-Xenium DAPI images were aligned with the spatial transcriptomic sample coordinates using 10X Xenium Explorer software (v.3.2.0). The coordinate-transformed mCherry immunofluorescence images were used as input to the Giotto calculate OverlapPolygonImages method to extract pixel intensities corresponding to the area occupied by each cell. Pixel intensities range was [0,255]. The background pixel intensities for each cell were extracted from a square region 60 mm across (±30 mm from cell centroid position) but excluding the cell itself. The cell mCherry signal enrichment score (RES) was computed as
$$\begin{array}{c}{\rm{RES}}({\rm{cell}})=({\rm{mean}}\,{\rm{cell}}\,{\rm{pixel}}\,{\rm{intensity}}+1)/\\ \,({\rm{mean}}\,{\rm{cell}}\,{\rm{background}}\,{\rm{pixel}}\,{\rm{intensity}}+1)\end{array}$$
The RES values were further converted to z-scores followed by thresholding, with mCherry+ cells determined by z ≥ 2 and mCherry− cells determined by z < 0.5. The mCherry+ set contained 401 cells (305 neurons) and the mCherry− set contained 19,042 cells (10753 neurons). Genes that were differentially expressed between mCherry+ and mCherry− cells within each neuronal cluster were identified using the scran method (FDR ≤ 0.1, and log2(fold change) ≥ 0.5). Global mCherry+ versus mCherry− differential genes were identified via the Nebula59 R package on all neurons simultaneously, by using the neuronal cluster assignment as a covariate in the model. Significant DEGs were selected as FDR < = 0.01 and log2(fold change mCherry) ≥ 0.3.
Quantification and statistical analysis
All data are presented as mean ± s.e.m. unless otherwise indicated. No statistical methods were used to pre-determine sample sizes, but our sample sizes are similar to those reported in previous publications23,60. Where possible, in vivo data were collected and analysed by an investigator blinded to the treatment group, but this was not always possible given the familiarity with the subjects. Determining whether injection sites were valid for inclusion or exclusion was determined by an investigator blinded to the results for that mouse. For histology, hormone, protein and RNA and metabolomic analyses, investigators were blinded to the group or treatment until data processing for group comparisons.
Injection sites were visualized and verified following behavioural experiments. Mice were excluded for virus expression outside of the MeA or for insufficient virus expression within the MeA. All mice in the Cre-independent DREADD activation experiment (Fig. 1k–o, Extended Data Fig. 2g–n and Supplementary Fig. 3a–j) showed viral spread into the lateral hypothalamus; data shown are from DREADD mice with >60% virus expression in the MeA.
Analyses were performed in RStudio or with Prism (v.10.3.1, Graphpad). Analyses in R were performed with R 3.6 using the lme4, lmerTest, emmeans, and car packages61,62. If the total number of data points for an experiment was less than 30, the data were tested for normality using the Shapiro–Wilk test. If the data were normally distributed or n > 30, data were analysed using Student’s unpaired two-tailed t-test for comparison between 2 groups, and one-way ANOVA with Tukey’s post hoc honest significant difference for comparison between multiple groups. Repeated studies were examined using a generalized linear mixed model with mouse identity as a random effect to account for repeated sampling across time or two-way repeated measures ANOVA. Cohort was included as a fixed variable where applicable. P values were adjusted using post hoc testing (for example, Tukey or Sidak’s testing) for multiple comparisons. If the data were not normally distributed, they were analysed with the Mann–Whitney U test or Kruskall–Wallis rank sum test with Dunn’s post hoc tests. Outliers were defined as values two standard deviations above or below the mean per group per time point (where applicable) and removed from analyses. P values below 0.05 were considered to be significant.
Figure panels were generated with Biorender (https://app.biorender.com/) and in Adobe Illustrator (v.2020).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.