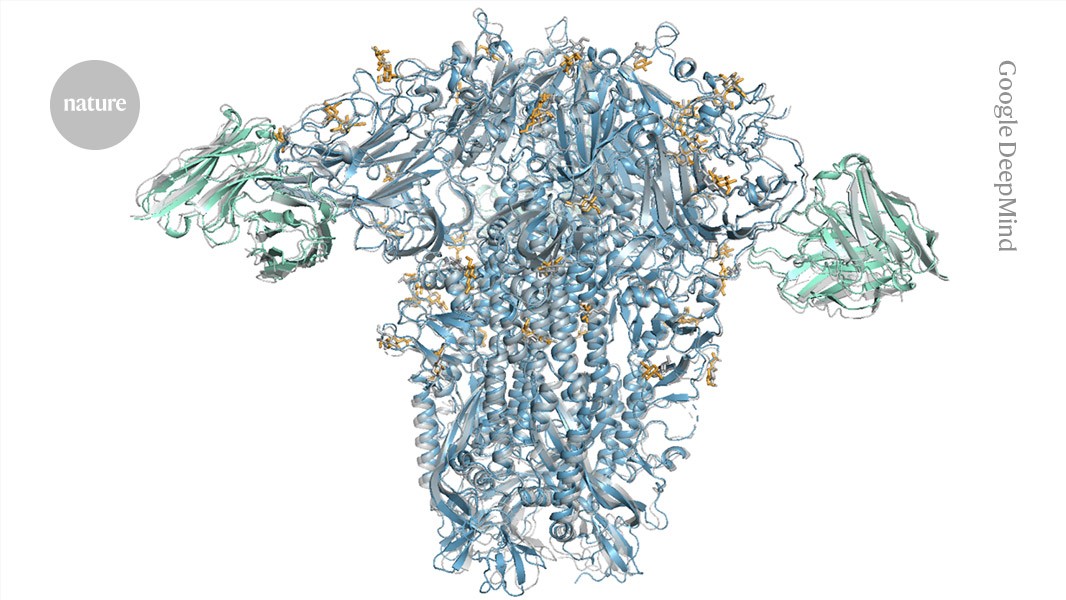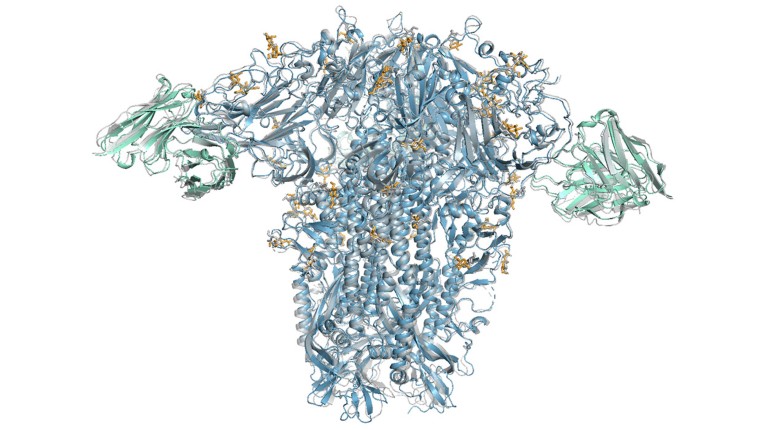
An AlphaFold 3 model of a common cold spike protein (blue) interacting with antibodies (green).Credit: Google DeepMind
AlphaFold, the revolutionary, Nobel prize-winning tool for predicting protein structures, has a problem: it’s running low on data.
The latest version of the artificial intelligence (AI) model, AlphaFold 3, has been touted as a game-changer for drug discovery, because it can model the interaction of proteins with other molecules, including drugs.
But a lack of examples of these interactions in the data underpinning AlphaFold — hundreds of thousands of publicly available protein structures — is holding the tool back for the applications that drug companies are most interested in, say scientists.
AlphaFold touted as next big thing for drug discovery — but is it?
A consortium of leading pharmaceutical companies announced plans today to make their own AlphaFold-3-inspired AI model using thousands of protein structures that are currently secreted away in company vaults. This is in addition to the more than 200,000 protein structures freely available in the Protein Data Bank (PDB).
“The data that’s missing from the PDB is exactly the data that’s present in our internal data,” says John Karanicolas, head of computational drug discovery at the pharma company AbbVie in Chicago, Illinois, and part of the effort, called the AI Structural Biology Consortium.
The consortium’s model will be based on OpenFold 3, a fully open-source reproduction of AlphaFold 3 that has been developed by academic researchers (using only publicly available data) and is due to be released in April. But there are no plans to make the consortium’s model available beyond member companies, which include AbbVie, Johnson & Johnson, Sanofi and Boehringer Ingelheim.
Google DeepMind, the London-based company that developed AlphaFold, is not involved in the project and did not wish to comment. Its spin-off company, Isomorphic Labs, is using AlphaFold 3 as part of collaborations with drug companies, including Novartis and Eli Lilly.
Drug data shortage
AlphaFold’s ability to predict proteins’ 3D shapes from their sequences relies on access to the PDB’s huge collection of protein structures mapped with experimental methods, such as X-ray crystallography. Many of these structures include interacting molecules — but they tend to involve biological partners such as the cellular energy source ATP, rather than drug compounds, says Karanicolas.
As a result, AlphaFold 3 does an adequate job of predicting how proteins interact with would-be drugs, but “it’s still a very open problem”, says Mohammed AlQuraishi, a computational biologist at Columbia University in New York City who is leading the development of OpenFold.
It’s possible that pharma-company protein structures, which are rarely deposited in the PDB, could help. As part of drug-development campaigns, firms routinely determine multiple structures for the same protein bound to many different drug candidates.
AlphaFold’s new rival? Meta AI predicts shape of 600 million proteins
The full extent of these proprietary protein-structure data isn’t known. But the data could equal or even exceed those of the PDB, says Stephen K. Burley, a director of one of the organizations that hosts the repository and a structural biologist at Rutgers University in Piscataway, New Jersey. AbbVie alone is contributing more than 9,000 structures to the consortium’s AI model. “It’s kind of crazy how much data there is sitting behind these walled gardens,” says AlQuraishi.
Drug companies won’t be sharing actual protein structures with each other — or with AlQuraishi — to develop the new model. Instead, the effort will use a platform developed by Apheris, a Berlin-based start-up company, that will allow OpenFold 3 to be retrained using proprietary data and without the structures ever leaving a company’s digital walls. Karanicolas says it will not be possible to reverse engineer the model to identify the secret structures it was trained on.
Whether the extra data will boost AlphaFold’s ability to model how proteins and drugs interact is unclear, says AlQuraishi. “That’s going to be the key question — what will the gains look like?” His team will evaluate the model, for example by comparing its predictions with experimental results, and make a detailed analysis public.
“I do think the experiment, negative or positive, is incredibly valuable,” he says. Some scientists and funding agencies are looking to create structural databases like those of the pharma companies with which to feed AI models, says AlQuraishi, and it will be worth knowing whether having more data is actually useful.