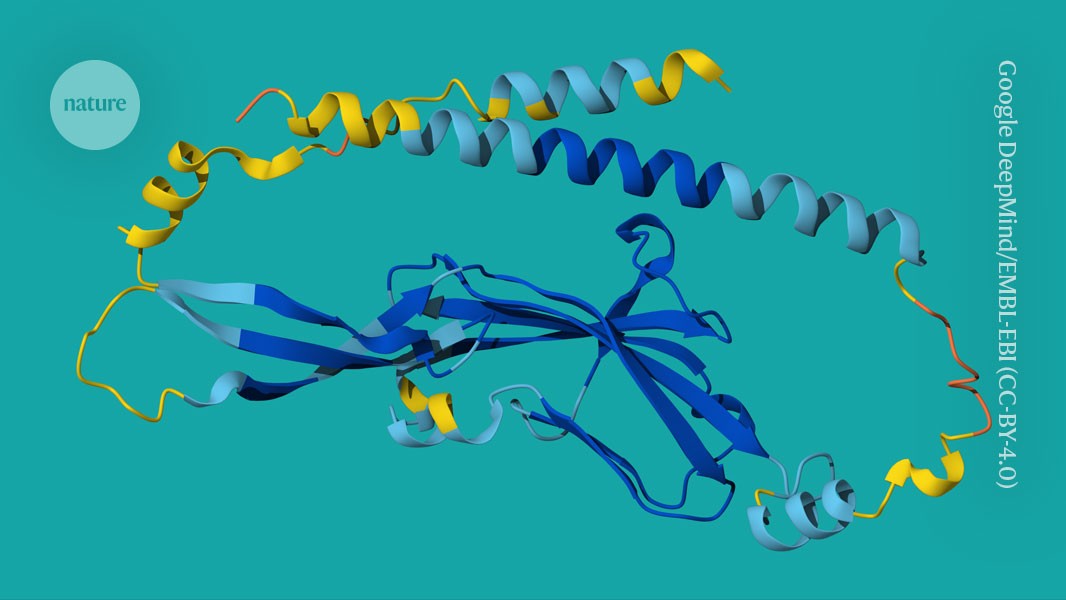
An AlphaFold model of Tmem81, a membrane protein involved in the fusion of egg and sperm.Credit: Google DeepMind/EMBL-EBI (CC-BY-4.0)
For nearly a decade, Andrea Pauli, a biochemist at the Research Institute of Molecular Pathology in Vienna, has been trying to work out how sperm and egg get together.
In 2018, her laboratory found a protein on the surface of zebrafish (Danio rerio) eggs, called Bouncer, that was essential for fertilization. But Pauli’s team and others struggled to show how Bouncer recognized sperm cells. Then a revolution happened.
AlphaFold reveals how sperm and egg hook up in intimate detail
Five years ago, in late November 2020, researchers at London-based Google DeepMind unveiled AlphaFold2. The artificial intelligence tool for predicting protein structures generated stunningly accurate 3D models that, in some cases, were indistinguishable from experimental maps, dominating a long-running structure-prediction challenge. The first version of AlphaFold was announced in 2018, but its predictions weren’t nearly as good as its successor, which limited its impact.
The 2021 release of AlphaFold2’s code and a database that has swelled to hundreds of millions of predicted structures mean that scientists can now get a reliable prediction for almost any protein.
“Having models for anything has had a huge impact,” says Janet Thornton, a bioinformatician at the European Bioinformatics Institute in Hinxton, UK, part of the European Molecular Biology Laboratory (EMBL-EBI). “It’s like the second coming of structural biology.”
Rapid discovery
For Pauli’s team, the software shone a light on a path they might otherwise never have found. The model predicted that a protein, called Tmem81, stabilizes a complex of two other sperm proteins, creating a pocket for Bouncer to bind to1. Experiments backed up the tool’s predictions. AlphaFold “speeds up discovery”, says Pauli. “We use it for every project.”

Source: OpenAlex/Google DeepMind
Her team’s paper about this, published in 2024, is one of nearly 40,000 journal articles to cite the 2021 Nature paper describing AlphaFold22. Unlike many other highly cited life-sciences and biomedical papers from the same period, including seminal reports about the COVID-19 pandemic, interest in AlphaFold doesn’t seem to be slowing down (see ‘Peak citations’).
DeepMind’s John Jumper — who shared half of the 2024 Nobel Prize in Chemistry with chief executive Demis Hassabis for developing AlphaFold — says he is “deeply proud” of how useful the tool has been for scientists such as Pauli. “When will someone win one of these major awards because they used AlphaFold?” he wonders.
Part of AlphaFold2’s rapid impact is down to its accessibility, say researchers. Google DeepMind made the underlying code and other parameters freely available to scientists, and it quickly became possible for them to run the software themselves at scale: this is what Pauli’s team did.

Some 3.3 million users in more than 190 countries have accessed the AlphaFold database (AFDB), which is hosted by EMBL-EBI and contains more than 240 million structural predictions, encompassing most known proteins. More than one million AFDB users come from low- and middle-income countries, including China and India (see ‘Global appeal’).
Protein-structure revolution
The field in which AlphaFold seems to have made its biggest impact is structural biology. Researchers who used AlphaFold submitted around 50% more protein structures to a repository of experimental models, called the Protein Data Bank (PDB), than did a non-AlphaFold-using ‘baseline’ of structural-biology researchers, finds a Google DeepMind-funded study of AlphaFold’s impacts released this week. AlphaFold2 use was also associated with higher rates of PDB submissions than those of researchers using other ‘frontier’ methods in AI, structural biology and protein structure prediction (see ‘Protein pile-up’).

Source: AI in science: Emerging evidence of impact from AlphaFold2
Jumper says he is especially gratified that AlphaFold2 — which was trained using PDB data — has proved so useful for deducing protein structures. The predicted structures can help researchers to make sense of raw data generated by X-ray crystallography and cryo-electron microscopy. “I love that it helps the people that gave us the data,” Jumper adds.