Microorganisms and culture conditions
R. delemar (99-880)9, R. arrhizus (557969)48 and Cunninghamella bertholletiae (506313)48 have been previously described. Mucor circinelloides (JMRC:NRZ:0774), Rhizomucor pusillus (JMRC:NRZ:0496), Rhizopus microsporus (JMRC:NRZ:0680), A. fumigatus (ATCC, 46645), Aspergillus flavus (JMRC:NRZ:0756), Aspergillus terreus (JMRC:NRZ:0442), C. albicans (JMRC:NRZ:1000), C. glabrata (JMRC:NRZ:1006), C. albicans SC5314 and Fusarium proliferatum (JMRC:NRZ:0657) were obtained from the Leibniz Institute for Natural Product Research and Infection Biology, Hans Knöll Institute. All Mucorales isolates were cultured on potato dextrose agar (Becton Dickinson) plates for 7 days at 37 °C. R. delemar M16 is a previously described pyrF-null mutant derived from R. delemar 99-880 that is unable to synthetize uracil49. R. delemar transformed with RNAi targeting mucoricin expression and R. delemar transformed with empty plasmid (control) were derived from strain M16, as previously described9. For the experiments involving these RNAi strains, a synthetic medium containing yeast nitrogen base (YNB, BD) supplemented with a complete supplemental mixture without uracil (CSM−URA, MP Biochemicals) (that is, YNB + CSM−URA) was used.
All of the bacterial isolates used (Escherichia coli, Pseudomonas aeruginosa, Klebsiella pneumoniae and Staphylococcus aureus) were clinical isolates obtained from the University Hospital of Heraklion, Crete. All bacteria were streaked onto plates containing LB agar plates from a freshly prepared frozen glycerol stock. After growth on LB agar plates, single colonies were used to inoculate overnight cultures of LB (50 ml at 37 °C). The next day, a volume of 1 ml of each of these cultures was diluted 1:10 to a final volume of 10 ml in LB medium and incubated in conical flasks for 2 h, at 37 °C, with constant shaking, to reach mid-log phase of growth. A volume of 200 μl was taken from each culture and the optical density was measured at 600 nm (OD600). The desired volume from each culture was used and centrifuged at 2,000 rpm for 2–3 min. Bacteria pellets were washed three times with LB medium. Bacteria pellets were diluted in RPMI medium or RPMI plus 4.5 g dl−1 of albumin to the desired OD600 of 0.2. A volume of 200 μl from each test condition was transferred to a flat-bottom 96-well plate. The plates were incubated at 37 °C, with constant shaking. At regular time intervals (45 min), 10 μl from each culture condition was diluted in RPMI medium to a final volume of 100 μl and the OD600 was measured spectrophotometrically. The mean value of triplicate measurements of bacterial growth in regular medium at t = 340 min was defined as 100% growth.
For filamentous fungi, the effect of albumin on fungal growth was assessed by counting spore germination rates in RPMI medium compared with RPMI supplemented with 4.5 g dl−1 albumin at 6 h using light microscopy. At least 300 spores were counted per condition in biological triplicates. For the effect of human IgG (i.v. human IgG, Gaminex, 100 mg ml−1, Grifols) fungal spores were incubated in medium containing increasing concentrations of human IgG (from 1.5 g dl−1 to 4.5 g dl−1). At 6 h, spores were assessed by light microscopy and photos were taken.
For Candida isolates, the impact of albumin on growth was evaluated spectrophotometrically. C. albicans strain SC5314 and Candida glabrata (JMRC:NRZ:1006) were streaked from freshly prepared glycerol stocks onto Sabouraud dextrose agar plates and incubated overnight at 37 °C. Single colonies were used to inoculate Sabouraud dextrose broth (50 ml) and liquid cultures were grown overnight at 37 °C with agitation at 150 rpm. The OD600 of the overnight cultures was determined, and aliquots were diluted with phosphate-buffered saline (PBS) to an OD600 of 0.1. Cells were collected by centrifugation at 2,000 rpm for 10 min, washed three times with PBS, counted and resuspended in either RPMI medium alone or RPMI supplemented with 4.5 g dl−1 albumin to an OD600 of 0.1. For each condition, 200 μl of the cell suspension was transferred into wells of a flat-bottom 96-well plate and incubated at 37 °C with continuous shaking. After 8 h, 10 μl from each well was diluted in RPMI to a final volume of 100 μl, and the OD600 was measured spectrophotometrically. Growth in standard RPMI medium was assessed in triplicate, and the mean OD600 value at 8 h was defined as 100% growth for normalization.
Human albumin depletion and purification
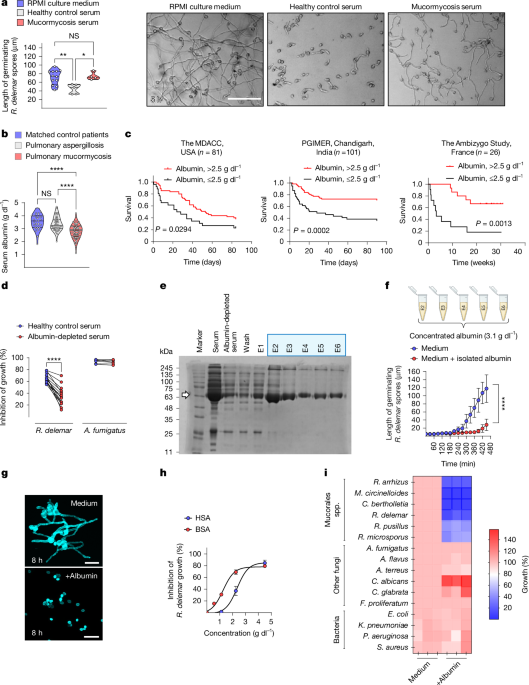
Albumin was depleted from human serum using an Albumin Depletion Kit (Abcam) according to the manufacturer’s instructions with a modification in the rehydration step of Cibacron Blue 3G-A beads, which was performed using albumin-free serum filtrate (generated through serum filtration using Amicon 50 kDa molecular weight cut-off (MWCO) ultracentrifugal filters; Merck).
For HSA purification, Blue Sepharose 6 Fast Flow (GE Healthcare) was first rehydrated with an albumin-free serum filtrate, incubated with fresh human serum (3 ml) at 4 °C overnight with rotation and then packed back in a column. The first volume (3 ml) of the flow-through contained albumin-depleted human serum. The column was subsequently washed with 7 ml of wash buffer (20 mM Na2HPO4, 20 mM NaH2PO4, pH 8). Albumin was isolated from the column in six consecutive elutions using 7 ml of elution buffer (2 M NaCl, 20 mM Na2HPO4, pH 8) each time. Elution fractions 2–6 were pooled and further processed for in vitro experiments.
Owing to the increased amount of NaCl in the eluted fractions, dialysis was performed using a CelluSep T2 membrane (Orange Scientific, Cellusep T2 Tubings, 6,000–8,000 MWCO), which was embedded in the appropriate buffer (20 mM Na2HPO4, 20 mM NaH2PO4, pH 8) for 4 h at 4 °C to achieve a physiologically relevant NaCl concentration (150 mM). Elutions were then condensed using Amicon 3 kDa MWCO ultracentrifugal filters (Merck), to a final volume of 2 ml and filter-sterilized through 0.22 μm Spin-X centrifuge tube filters (Costar). The flow-through generated during the condensation procedure was pooled, measured for any remnants of albumin (no traces of albumin were found) and also filter-sterilized using the same 0.22-μm filters.
Chemical modifications of albumin
Highly oxidized albumin was prepared by the incorporation of cysteine into reduced albumin50. BSA was treated with a 50-fold molar excess of L-cysteine/cystine by mixing 80 ml of 0.06 mM BSA with 72 ml of 3 mM cysteine and 8 ml of 3 mM cystine. All of the solutions were diluted in 0.1 M sodium carbonate and hydrogen carbonate buffer (pH 10) and, after incubation at 37 °C for 48 h, the resulting mixture was lyophilized. Next, the residue was dissolved in PBS and purified from low-molecular-mass components (excess cysteine/cystine) by filtration through an Ultrafree-3000 Da membrane (Millipore) at 4 °C. Purified, highly oxidized albumin was lyophilized and stored at room temperature. Glycosylated albumin was prepared by diluting albumin in PBS containing 5 mM glucose51. The solution was incubated for 72 h under an atmosphere of 95% O2, and 5% CO2 to maintain the pH at 7.3–7.4. The mixture was then lyophilized, followed by solubilization in PBS and purification through ultrafiltration with an Ultrafree-3000 Da membrane. Purified, glycosylated albumin was lyophilized and stored at room temperature. The control solutions were prepared in PBS and subjected to the same lyophilization and filtration processes to increase the reliability of the results. The dried, equal-weight samples were derivatized with 20 μl of 20 mg ml−1 methoxy amine/pyridine and 50 µl of hexane. The mixture was vortexed and kept at 37 °C for 1.5 h, with vortexing every 30 min. Then, 90 μl (N-trimethylsilyl-N-methyl trifluoro-acetamide and trimethyl-chlorosilane (MSTFA + 1% TMS) were added and the samples were vortexed for 30 s and incubated at 37 °C for 1 h. The obtained solution was filtered through 0.45-μm syringe filters (nylon syringe filter, Membrane Solutions) and subjected to GC–MS/MS in multiple reaction monitoring (MRM) mode analysis according to previously published methods52,53. The obtained metabolites were imported into MetaboAnalyst (v.5.0; https://www.metaboanalyst.ca/home.xhtml). Hierarchical cluster analysis and partial least squares-discriminant analysis were then performed according to previous methods54.
The removal of FFAs from albumin (BSA and HSA isolated from donors) was performed with the use of activated charcoal, as previously described with some modifications25. Next, activated charcoal and albumin were mixed at a ratio of 1:2 for 1 h at 4 °C. The excess of activated charcoal was removed by ultracentrifugation at 20,200g for 30 min at 4 °C.
Loading of albumin with FFAs was performed as previously described55,56 with some modifications. In brief, 4.5 g dl−1 charcoal-treated albumin (diluted in RPMI medium) and 80 mM FFAs (diluted in ethanol) were heated at 55 °C for 30 min and 5 min, respectively. Then, FFAs and albumin were gently mixed at a ratio of 8:1. The FFA–albumin mixture was thoroughly mixed by vortexing and incubated at 37 °C for 1 h. Effective conjugation of FFAs to albumin resulted in a clear solution.
Extraction and quantification of caprylic acid in glycosylated and non-glycosylated BSA
Quantification of caprylic acid in glycosylated and non-glycosylated BSA was conducted by extracting caprylic acid using chloroform/methanol solvent (2:1, v/v)57. This was followed by centrifugation at 3,500 rpm to separate the organic layer, which was filtered through anhydrous sodium sulphate and then dried using a rotary evaporator (Büchi). The dried residue was subsequently derivatized and analysed using GC–MS/MS in MRM mode against standard caprylic acid (m/z 201.10, 117.10 and 75.00).
Fluorescence displacement assay of caprylic acid binding with glycosylated and non-glycosylated BSA
The fatty-acid binding affinity of glycosylated and non-glycosylated BSA was assessed using the 1-anilino-8-naphthalene sulfonic acid (ANS) fluorescence displacement assay according to a previously established protocol58. In brief, BSA solutions (2.5 μg in 100 μl PBS) were added into black 96-well flat-bottom microplates (Nunc Microwell Plates), followed by the addition of 50 μl ANS in PBS to a final concentration of (0, 1.25, 2.5, 5 and 10 μM) with incubation at room temperature in dark for 15 min. For the displacement assay, caprylic acid was added at increasing concentrations (0–10 μM) to the ANS–BSA complexes. A control without caprylic acid was included to assess the initial fluorescence. The mixtures were incubated at room temperature in the dark for an additional 15 min. Measurements were performed using a microplate reader (Synergy H1, Biotek) with excitation and emission wavelengths set at 360 nm and 460 nm, respectively. Fluorescence quenching, indicative of ANS displacement by caprylic acid, was recorded as the percentage change in fluorescence intensity (ΔF%) using the formula: %ΔF quenching = [(F0 − F1)/F0] × 100, where F0 is the fluorescence in the absence of caprylic acid and F1 is the fluorescence at each tested concentration of caprylic acid.
In vitro assessment of the inhibitory activity of albumin and human serum
Fungal conidia (spores) were collected by gentle shaking in the presence of sterile 0.1% Tween-20 in PBS, washed twice with PBS, filtered through a 40-μm-pore-size cell strainer (Falcon) to separate conidia from contaminating mycelium, counted using a haemocytometer and suspended at a concentration of 108 spores per ml. To achieve synchronized swelling of Rhizopus conidia, 1 × 106 per ml dormant conidia were incubated for 4–6 h at 28 °C in a six-well plate in RPMI-MOPS (pH 7.0) supplemented with 2% glucose with or without 4.5 g dl−1 BSA or other sources of albumin. To study the minimal growth requirements of R. delemar, a minimal-growth-requirements medium was used59 based on HBSS supplemented with 0.05% (w/v) MgSO4·7H2O, 0.1% (w/v) glucose and 0.1% (w/v) NH4Cl (minimal medium). For fluorescence labelling, the spores were stained with 20 μg ml−1 Fluorescent Brightener 28 (Calcofluor White; CFW, Sigma-Aldrich) in 0.1 M NaHCO3 pH 8.3 at room temperature in the dark for 1 h, with constant rotation.
In another set of experiments, R. delemar spores were incubated at 37 °C in RPMI-MOPS without phenol red adjusted to pH 7, for 2 h to start swelling. Then, 3.6 g dl−1 BSA and 900 μg ml−1 FITC-albumin (Sigma-Aldrich) were added to the medium and the spores were cultured in a 96-well μ-Plate (Ibidi) at 37 °C. After 5 h of culture, R. delemar live spores were collected, washed twice with PBS and then stained with 100 μg ml−1 CFW in 0.1 M NaHCO3 pH 8.3 at room temperature in the dark for 1 h, with constant rotation. Fungal cells were then imaged with a spinning-disk confocal microscope (Dragonfly 200, Andor).
To assess the inhibitory activity of human serum, 1 × 104 fungal spores were incubated in flat-bottom 96-well plates containing 100 μl of serum from healthy individuals or patients at 37 °C and 5% CO2 for 5–6 h. At least three different fields of each well (a total of >100 germinating spores) were imaged under an inverted microscope (Olympus) and the length of the germinating conidia was measured using ImageJ60. In certain experiments, time-lapse videos of fungal growth were acquired using an Operetta high screening content system (Perkin Elmer) at room temperature with 5% CO2. Photographs of each well were automatically taken approximately every 30 min for 18 timepoints and analysed using Harmony v.4.1 software (Perkin Elmer).
The inhibition of fungal growth was additionally quantitated via an XTT metabolic activity assay (Biotium). A solution of XTT tetrazolium salt (0.25 mg ml−1) and menadione (25 μM) was freshly prepared in PBS and added to the cell culture. Fungal spores were further incubated at 37 °C for another 1 h (ref. 61). Fungal-spore-free culture supernatants were collected, and their absorbances at 450 nm (OD450) and 620 nm (OD620) were measured using a microplate photometer (Multiscan, Thermo Fisher Scientific). Growth inhibition was calculated according to the following formula:
$$\text{Growth inhibition}({\rm{ \% }})=100\times \frac{({{\rm{O}}{\rm{D}}}_{450}-{{\rm{O}}{\rm{D}}}_{620}{)}_{\text{control cells}}-({{\rm{O}}{\rm{D}}}_{450}-{{\rm{O}}{\rm{D}}}_{620}{)}_{\text{treated cells}}}{({{\rm{O}}{\rm{D}}}_{450}-{{\rm{O}}{\rm{D}}}_{620}{)}_{\text{control cells}}}$$
In vitro studies on the antimicrobial activity of FFAs
FFA stock solutions were initially dissolved in absolute ethanol to a final concentration of 80–1,000 mM and incubated at 95 °C for 5 min. 2× MOPS-RPMI medium was also heated at 95 °C for 30 min and serial dilutions of FFAs were made by slowly adding the desired volume of preheated FFAs into the medium. The final concentration of ethanol used under culture conditions was 5% to ensure optimal dilution of FFAs; we confirmed that 5% ethanol had no significant inhibitory effect on Mucorales growth. Fungal spores were incubated in FFA-containing MOPS-RPMI medium at 37 °C for 16–18 h and fungal germination was measured as previously described13.
Lipid extraction
Lipids were extracted using a CHCl3:CH3OH (2:1) solution according to a previously published method62, dried with anhydrous sodium sulfate, subjected to residual evaporation via a rotatory evaporator (Buchi) under an inert stream of nitrogen and then stored at −20 °C (ref. 63). The residue obtained was equally divided into two parts; one for the oxidation reaction and the other left under inert conditions to avoid auto-oxidation.
Microwave-assisted oxidation of FFAs
FFAs and BSA-bound FFAs were subjected to oxidation through an irradiation microwave reactor according to modified previously published methods64. The samples were dissolved in 0.2 mM H2O2 in methanol to a final volume of 10 ml. The oxidization reaction was performed in a sealed vial on a synergy microwave synthesizer (CEM) for 20 min at an operating temperature of 100 °C and a pressure of 200 psi. The reaction mixture was extracted with CHCl3, dried with anhydrous sodium sulfate, subjected to residual evaporation through a rotatory evaporator (Buchi) and stored at −20 °C.
Open-air-assisted oxidation of FFAs
FFAs (OA), human or mouse plasma lipid extracts were diluted in absolute ethanol and exposed to open air inside a laminar flow hood as previously described65. After 4 days, ethanol was evaporated, and the precipitates of the oxidized FFAs were rediluted to their initial volume in 0.01 M Tris-HCl pH 7.5 (human and mouse plasma lipids) or in absolute ethanol to a final concentration of 80 mM (OA) and stored at −20 °C.
Cell damage assay for the assessment of toxin neutralizing activity of albumin
Human alveolar epithelial cells (A549 cells) were obtained from a 58-year-old male Caucasian patient with carcinoma and procured from American Type Culture Collection (ATCC). The cells were propagated in F-12K Medium developed for lung A549 epithelial cells. The damage of A549 epithelial cells was quantified using a 51Cr-release assay9,66. In brief, confluent cells grown in 24-well tissue culture plates were incubated with 1 μCi per well Na251CrO4 (ICN) in F12K-medium for 16 h. On the day of the experiment, the medium was aspirated, and cells were washed twice with pre-warmed Hanks’ balanced salt solution (HBSS, ScienCell). Cells treated with mucoricin with or without the addition of albumin (free of FFAs BSA at a final concentration of 4.5 g dl−1) were suspended in 1 ml of F12K-medium supplemented with glutamine and incubated at 37 °C in a 5% CO2 incubator. Spontaneous 51Cr release was determined by incubating the untreated cells in the same volume of the culture medium supplemented with glutamine. At different timepoints, and after data were corrected for variations in the amount of tracer incorporated in each well, the percentage of specific cell release of 51Cr was calculated as follows: [(experimental release) − (spontaneous release)]/[1 − (spontaneous release)]66. Each experimental condition was tested at least in triplicate, and the experiment was repeated at least twice.
Ethics statement on human clinical studies
All research involving human participants was conducted in accordance with relevant ethical regulations and approved by the ethics committees of the respective participating institutions. All patients or their legal representative provided written informed consent for the collection of clinical samples analysed in this study, in accordance with the Declaration of Helsinki.
The medical records of contemporaneous patients who had been admitted at the Leukemia Department of the MD Anderson Cancer Center were retrospectively reviewed. Standardized EORTC/MSG criteria were applied for diagnosis of pulmonary mucormycosis and pulmonary aspergillosis67. Clinical information of matched control patients for the underlying disease who developed bacterial (Legionella) pneumonia were also reviewed. Albumin levels were retrieved from the medical records of all of the patients on the day of hospital admission. Information on demographics, underlying disease and risk factors for mucormycosis were collected. The study was approved by the Institutional Review Board (IRB) of The University of Texas MD Anderson Cancer Center (MDACC; protocol PA14-0802).
The medical records of consecutive patients admitted to the Department of Pulmonary Medicine, Department of Medical Microbiology, Institute of Medical Education and Research (PGIMER), Sector-12, Chandigarh, 160012, India over the past 8 years (2016–2023) with a diagnosis of pulmonary mucormycosis were retrospectively evaluated. The study was approved by the Institutional Ethics Committee (IEC; Intramural; INT) of the PGIMER (IEC/INT, 2023/study-1564) and registered with the Biological Research Regulatory Approval Portal (BIORRAP, POS36452025). All procedures were conducted in accordance with institutional and national guidelines and regulations.
Serum samples were prospectively collected from healthy individuals, patients with cirrhosis and patients with haematological malignancies at the University Hospital of Heraklion, Crete, and the Therapeutics Clinic of the General Hospital of Athens “Alexandra”. Demographics, clinical characteristics, serum albumin levels, total FFAs and oxidized FFA levels were measured in the sera of healthy individuals and patients. Approval for the collection of clinical information and blood samples from all individuals mentioned above was obtained from the Ethics Committee of the University of Heraklion, Crete, Greece (5159/2014, 10925/201 and 13-04-22/7970) and Therapeutics Clinic (122/18-2-2021).
For lipidomic and functional studies, serum samples from patients with mucormycosis or invasive pulmonary aspergillosis, and from matched control patients were obtained from patients with haematological malignancy admitted to the University Hospitals of Leuven, Belgium (IRB, S61797).
Vitamin quantification in culture medium using LC–MS
Samples were first filtered through 0.22-μm nylon syringe filters before undergoing LC–MS/MS analysis. The analysis was performed via an Elute UHPLC system linked to a Q-TOF mass spectrometer. Chromatography separation was performed on a Hamilton Intensity Solo C18 column, maintained at 35 °C. The sample mixture was 10 μl in a solvent system composed of 0.1% formic acid in deionized water (solvent A) and acetonitrile (solvent B). The elution gradient progressed from 1% to 99% solvent B over 20 min, with flow rates adjusted between 0.25 ml min−1 and 0.35 ml min−1. Vitamin detection followed established methods by employing standards of cyanocobalamin (B12), folic acid (B9), riboflavin (B2), thiamine (B1), biotin (B7), Ca-panthenoate (B5), pyridoxine (B6), nicotinamide (B3), choline chloride and PABA, all sourced from Sigma-Aldrich. Stock solutions of 5 mM for vitamins B1, B3, B5, B6, B7, B9 and B12, and choline chloride were prepared in water, with riboflavin (B2) in DMSO. Internal standard solutions were made at 10 mM, and working solutions comprised 200 mM of all vitamins in water and 100 mM of internal standards. Calibration curves were prepared using 0.1% formic acid in water, spanning six serial dilutions from 0 to 100 mM, each including 2.5 mM internal standards. Samples and calibration solutions underwent identical processing involving liquid-liquid extraction and drying before analysis, ensuring precise quantification of vitamins with detection limits in the low µg l−1 range. Each sample was injected three times.
Analysis of trace elements in culture medium using ICP-MS
Inductive coupled plasma MS (ICP-MS-9800 Series) was used to detect trace metals in the samples. The samples were collected and digested with hydrochloric acid, to ensure complete dissolution. Microwave-assisted digestion was used for rapid and efficient sample preparation. The digested samples were then diluted with deionized water such that the concentration was within the measurable range of the ICP-MS instrument. Calibration standards with known concentrations of Na, K, Ca, Mg, Cu and Fe were prepared, and isotope-labelled internal standards were used to correct for matrix effects and signal fluctuations. The prepared samples were injected into the ICP-MS instrument through a nebulizer, which converts the liquid sample into an aerosol. The ionized elements emit light at characteristic wavelengths, which are detected and quantified by a mass spectrometer. The concentrations of trace elements in the samples were calculated by comparing the sample signals to the calibration curve.
Analysis of amino acids in culture medium
The samples were centrifuged at 12,000 rpm for 10 min and the supernatants were analysed using the Waters Acquity ultra-performance liquid chromatography (UPLC) H-Class-Xevo TQD system equipped with ESI. HPLC was performed using the Acquity BEH C18 column (1.7 μm, 2.1 mm × 150 mm). The binary mobile phase consisted of solvent A (100% methanol) and solvent B (0.2% formic acid). The column gradient was as follows: 100% B for 10 min; 66.6% B for 0.5 min; returned to 100% B for 1 min; and maintained at 100% B for another 0.5 min. The flow rate was 0.2 ml min−1, the injection volume was 10 μl and the column effluent was monitored by MS. The MS analysis was performed in MRM mode in positive-ionization mode. The MS parameters were as follows: the dwell time was 0.02 s, and nitrogen was used as a desolvating gas at a flow rate of 600 l h−1. The ionization source conditions were as follows: desolvation temperature 350 °C; source temperature 150 °C; collision gas (argon) flow 0.1 ml min−1; and capillary voltage 3.0 kV. The parameters of mass analyser were set as follows: the LM1 and HM1 resolutions were 15 and 15, respectively, as ion energy 1; the LM2 and HM2 resolutions were 15 and 15, respectively, and the ion energy was 2. The amino acids in the samples were identified on the basis of the mass-to-charge ratio.
The quantification of amino acids was conducted using a standard mix calibration curve, with the UPLC–MS/MS system controlled by Lynx software (v.4.1, SCN 882). Data analysis was performed via the TargetLynx program. Each sample, which contained a mixture of amino acids at specific concentrations, was injected three times to ensure accuracy. The amino acid standard solution (AAS18, Sigma-Aldrich) included 2.5 μmol ml−1 of various amino acids such as L-alanine, L-arginine and L-valine, with L-cystine at 1.25 μmol ml−1. The samples were diluted in 0.1% formic acid in water for analysis. Using the MIDAS workflow, full scan linear ion trap MS/MS data confirmed the identity of the target analytes. MRM extracted ion chromatograms were used for all amino acids, especially at concentrations near their limits of detection, ensuring precise quantification and linearity. Limits of detection were accurately calculated for all amino acids, and their concentrations were measured.
Albumin ligand analysis and fractionation
BSA was added to minimal growth requirements medium to a final concentration of 4.5 g dl−1, left at room temperature for 0.5–1 h and then filtered through Amicon 3 kDa MWCO ultracentrifugal filters (Merck) to remove albumin. BSA after filtration (15 ml) was subjected to fractionation through C-18 solid-phase extraction. After conditioning with 6 ml of methanol and 6 ml of ultrapure water, 15 ml of the sample was applied, giving fraction 1 (F1). Subsequently, with gradual elution (6 ml of 5% methanol), fraction 2 (F2) was obtained. Finally, fractions 3 (F3) and 4 (F4) were obtained using 6 ml of 50% methanol and 6 ml of pure methanol, respectively. During the entire process, the flow rate was held constant at approximately 3 ml min−1. The solvents and reagents used were of LC-MS grade (Merck).
For fraction component identification through GC-MS analysis, the obtained fractions (1–4) were derivatized with methyl-chloroformate (Sigma-Aldrich). Specifically, 300 μl of every fraction was mixed with 80 μl of pyridine, 200 μl of methanol and 50 μl of methyl-chloroformate, vortexed for 30 s and incubated for 6 min at room temperature. After incubation, 1 ml of hexane was added to each reaction mixture and extracted via the liquid-liquid extraction technique. Finally, the hexane phase was subjected to GC-MS analysis.
Fractions F1–F4 were analysed through gas chromatography coupled with a single quadrupole MS (GC-MS) analyser. GC-MS analysis was performed with an Agilent gas chromatography instrument (model 8860) system coupled to a mass spectrometer (model 5970) using an electron ionization (EI) source.
The samples were qualitatively analysed and caprylic acid (or octanoic acid) was identified (NIST 14.0 EI spectral library) as the main component of F2. Furthermore, the caprylic acid structure was verified using a reference standard (Sigma-Aldrich), and its concentration was quantitatively determined in all four fractions, following the same derivatization protocol and a standard solution calibration curve. The entire analysis was conducted in duplicate. F2 was the richest in caprylic acid, with an average concentration of 194.1 μg ml−1, followed by F3, with a much lower concentration of 15.2 μg ml−1. In F1 and F4, only traces of caprylic acid were detected.
F2 was also analysed using HRMS without any derivatization steps to verify the structure of caprylic acid. Specifically, an UPLC system coupled with a HRMS analyser, especially a triple-TOF 5600+ (AB SCIEX) system, was used in negative ESI mode. LC analysis was performed on the ACQUITY H-Class UPLC system (Waters) equipped with a binary solvent manager and an FTN sample manager. A volume of 10 μl of F2 was injected into the system and separated by a linear gradient containing water and acetonitrile (mobile phases A and B, respectively, 5–100%), with the aqueous phase containing 0.1% formic acid. The solvents were of LC-MS grade and purchased from Merck. The analysis was performed with a Fortis Speedcore Biphenyl reversed-phase chromatography column (2.6 µm, 2.1 mm × 100 mm). The Triple-TOF platform was equipped with a DuoSpray ion source. The acquisition mode used was data dependent, covering a mass range of 80–500 m/z. Specifically, the tripleTOF parameters for acquisition were as follows: source temperature of 450 °C, source voltage of −4,500 V, exhaust gas pressure of 50 psi and curtain gas pressure of 35 psi. The caprylic acid structure was verified at m/z 143.1079 [M-H]- a, with a mass error of 1.1 ppm and degree of unsaturation (rings and double bond equivalents) of 1 (Extended Data Fig. 4).
FFA extraction from human plasma
FFAs were extracted from 100 μl of previously thawed plasma by the addition of 300 μl of cold methanol/ethanol (1:1) (Thermo Fisher Scientific). C17-Sphinganine was added as an internal standard. The samples were vortexed for 1 min, incubated on ice for 5 min and centrifuged at 16,100g for 20 min at 4 °C, after which 100 μl of the supernatant was transferred into LC–MS vials for analysis.
FFA extraction from the plasma of albumin-KO mice
FFAs were extracted from 100 μl of previously thawed plasma by the addition of 300 μl of cold methanol (Thermo Fisher Scientific). Next, 10 μl of 2 μM 9(S)-HODE-d4 solution (Cayman Chemical) was added as an internal standard. The samples were vortexed for 2 min and then centrifuged at 16,100g for 10 min at 4 °C, after which 100 μl of the supernatant was transferred into LC-MS vials for analysis.
Non-targeted analysis by LC–MS for FFAs
LC–MS analysis was performed on a UHPLC system 1290 Infinity II (Agilent Technologies) coupled with a 6546 QTOF MS detector in negative ESI mode. For separation, a volume of 1 μl was injected onto a Zorbax Rapid Resolution High Definition Extend-C18 column (Agilent Technologies, 2.1 × 50 mm, 1.8 μm) thermostated at 60 °C. The flow rate was 0.6 ml min−1, with a mobile phase composed of ultrapure Milli-Q water with 0.1% formic acid for A and acetonitrile with 0.1% formic acid for B. The chromatography gradient started from 5% B for the first min and increased to 80% B in 6.0 min, then to 100% by 11.0 min, and the starting conditions were returned to 1.0 min, allowing re-equilibration until 15.0 min. Data were collected in negative ESI ionization mode and operated in the range of m/z 100–600 and m/z 40–600 for MS/MS analysis using iterative Agilent mode. The nozzle voltage was set to 1,000 V, and the capillary voltage was −4,000 V with a scan rate of 1.2 scans per s. The drying gas was heated to 250 °C at a rate of 12 l min−1 and a pressure of 52.0 psi. Additional heating was applied using sheath-heated gas up to 370 °C with a flow rate of 11 l min−1 to improve ionization. For internal mass correction during data acquisition, one reference mass was infused continuously into the system throughout the whole analysis: m/z 112.9856 (proton-abstracted TFA anion). An external calibration with FA 18:0 was used for semi-quantification of fatty acid species. Calibration curve samples were prepared at different concentrations (1.0, 2.5, 5.0, 10.0, 13.0, 17.0 and 20.0 ppm), and 9(S)-HODE-d4 was included as an internal standard at the same concentration as the serum samples. The raw data collected by LC-MS were reprocessed with MassHunter Profinder software v.B.10.02. Calibration curves were built using normalized values of FA 18:0 by d4-9-HODE versus concentration in ppm. The results are expressed as the concentration in mM of the corresponding fatty acid in the serum. Detailed methods for identification of serum oxylipins and the standards used for the analysis of FFAs are provided in the Supplementary Methods.
RNA isolation from R. delemar cells
At the indicated timepoints of incubation in medium with or without albumin (0 h, 3 h and 6 h), the R. delemar cells were removed by scraping, centrifuged at 400g and lysed with 450 μl of RLT buffer + β-mercaptoethanol using the RNeasy Plant Mini Kit (Qiagen). Each sample was subsequently sonicated using a sonication probe on ice for 20 × 1 s (set 40). RNA was then isolated according to the manufacturer’s instructions.
RNA-seq data generation and analysis
RNA-seq libraries (strand-specific, paired-end) were generated from total RNA by using a TruSeq RNA sample prep kit (Illumina). In total, 150 nucleotides of sequence were determined from both ends of each cDNA fragment using the HiSeq 4000 platform (Illumina). Sequencing reads were aligned to the reference genome (R. delemar 99-880) using STAR aligner (v.2.7.10)68. The reads that mapped to genomic features were calculated using the featureCounts program (v.2.0.3)69. Statistical analysis of differential gene expression was performed using the DEseq2 R-based package70. A gene was considered differentially expressed if the absolute log fold change was greater than or equal to 1 and the false-discovery rate value for differential expression was below 0.05. The RNA-seq analysis was performed in biological triplicate. RNA-seq data were generated by Maryland Genomics at the Institute for Genome Sciences, University of Maryland School of Medicine.
RNA-seq enrichment analysis
The GO enrichment analysis performed using the gene set enrichment analysis (GSEA)71 method as previously implemented by the web-based application FungiFun3 (https://fungifun3.hki-jena.de/)72. The BlastKOALA automatic annotation server73 was used for the functional annotation and the assignment of the KEGG orthologies. The pathway enrichment analysis performed using the generally applicable gene-set enrichment method74 and functions of the gage R-based package. The genes of the selected KEGG pathways were identified using the KEGG PATHWAY database. R programming language was used for the visualization of the results. The heat maps were produced using the pheatmap package75. All of the P values were adjusted using the Benjamini–Hochberg method and adjusted P < 0.05 was considered to be statistically significant.
Animal studies
B6.Cg-Tg(FCGRT)32Dcr Albem12MvwFcgrttm1Dcr/MvwJ mice, referred to as Alb−/− mice throughout (obtained from The Jackson Laboratory), and C57BL/6 mice were maintained in grouped cages in a high-efficiency air-filtered, environmentally controlled, virus-free facility (24 °C, 50–60% relative humidity, 12 h–12 h light–dark cycle) and fed a standard chow diet and water ad libitum.
Ethics statement on animal studies
All experiments were approved by the local Ethics Committee of the University of Crete Medical School, the FORTH Ethics Committee, and the Directorate of Agricultural and Veterinary Policy of the Region of Crete, in accordance with national and European Union legislation (animal protocols 17/07/2017-147075 and 22/03/2023-90477). All efforts were made to minimize the number of animals used and their suffering.
In vivo studies of fungal infection
For virulence studies, 8–12-week-old, age- and sex-matched mice were infected either through i.v. inoculation or through i.t. instillation of 1–5 × 106 spores of R. delemar or A. fumigatus, and their survival was monitored for 14–25 days. After infection, two mice from each group were euthanized for inoculum verification. For histopathological evaluation, lungs were collected at different timepoints: 0 h, 6 h and 1 day after infection. In a separate experiment, age- and sex-matched C57BL/6 mice were infected as above and then euthanized on day +1 after infection. Lungs were collected and processed to determine tissue fungal burden by qPCR. Approximately half of the lung tissue was snap frozen in ethanol/dry ice and stored at −80 °C until analysed76. In brief, lungs were homogenized in Whirl Pak bags (Thermo Fisher Scientific). Approximately 1.5 ml of the homogenate was transferred to sterile screw-cap lysing matrix tubes with 1.4-mm-diameter glass beads (MP Biomedicals). The homogenate containing mouse tissues and fungal hyphae was mechanically disrupted using Fastprep FP120 (Bio Thermo Electro Corporation) with three bursts of 30 s at speed 4 (with incubation on ice between bursts). The supernatant was collected from the secondary homogenate by centrifugation at 800g for 5 min at 4 °C. DNA was extracted from secondary homogenate with the DNeasy tissue kit (Qiagen) according to the manufacturer’s instructions. DNA was recovered in 200 μl of elution buffer and stored at 20 °C until analysis by qPCR. Oligonucleotide amplification primers of the R. oryzae 18S rRNA gene (GenBank: AF113440) were designed with Primer Express software (v.1.5; Applied Biosystems) and synthesized commercially (Sigma-Aldrich). The sequences of these oligonucleotides are as follows: (1) sense amplification primer, 5′-GCGGATCGCATGGCC-3′; and (ii) antisense amplification primer, 5′-CCATGATAGGGCAGAAAATCG-3′. The qPCRs were performed as described previously76. Five-point standard curves were prepared by spiking uninfected lungs with known concentrations of R. oryzae sporangiospores, extracting total lung DNA, and then analysing R. oryzae-specific nucleic acid concentrations by qPCR. The standard curve created by spiking spores was used to calculate log10-transformed spore equivalents per gram of tissue.
For the establishment of immunosuppression/neutropenia, 8–12-week-old age- and sex-matched mice were administered three intraperitoneal injections of cyclophosphamide (Sigma-Aldrich, 150 mg per kg body weight on days −4 and −1, 100 mg per kg on day +3) and a subcutaneous injection of cortisone acetate (Sigma-Aldrich, 300 mg per kg on day −1), as previously described77. The mice were then infected i.t. with 1 × 106 spores of R. delemar. For the prophylactic model, 50 mg of FFA-free HSA was administered intraperitoneally on days −6, −4 and −2 prior to pulmonary infection with R. delemar. For the pre-emptive therapeutic model, 25 mg of FFA-free HSA was administered intraperitoneally daily for 6 consecutive days starting 6 h after pulmonary infection with R. delemar.
For the model of disseminated candidiasis, the C. albicans strain SC5314 was subcultured twice at 37 °C for 48 h on Sabouraud dextrose agar. After the second subculture, single C. albicans colonies were placed in liquid Sabouraud dextrose medium and grown for 16–18 h in a shaking incubator at 30 °C and 150 rpm. Candida blastoconidia were collected, centrifugated at 2,000 rpm for 10 min, washed three times in PBS, counted and reconstituted in PBS to a final concentration of 5 × 106 cells per ml. Then, 150 μl of C. albicans cells diluted in PBS (7.5 × 105) were used to i.v. infect 10–12-week-old, age- and sex-matched, wild-type C57BL/6 and Alb−/− mice, as previously described78. The viability of the inoculated C. albicans blastoconidia was assessed by performing serial dilutions in PBS and plating onto Sabouraud dextrose agar. Plated C. albicans cells were incubated at 37 °C overnight and the next day the number of colonies was counted and the inoculum validated.
Histopathology and immunohistochemistry/immunofluorescence studies
The mice were euthanized, and their lungs were excised and fixed with 10% formalin before being embedded in paraffin and cut into 5–7 μm sections. Lung tissue sections were deparaffinized in xylene and rehydrated through an ethanol gradient (100–70%). H&E or GMS (Sigma-Aldrich) staining was performed for histopathology and fungal burden assessment, according to the manufacturer’s instructions. For histological quantification of fungal burden79, FFPE lung sections were stained with GMS to visualize fungal elements. In certain experiments, fungal conidia were counterstained with periodic acid-Schiff (PAS). Images were acquired from at least ten non-overlapping fields per section at ×10 magnification. Fungal hyphae were enumerated per high-power field in a blinded manner by an experienced pathologist.
For active caspase-3 immunostaining in the lung tissue, heat-induced antigen retrieval was performed to deparaffinize and rehydrate the lung sections through incubation in sodium citrate buffer (10 mM, 0.05% Tween-20, pH 6) for 40 min at 90–95 °C using a steamer (Philips). The tissue sections were allowed to cool for 30 min, washed three times in PBS and blocked endogenous peroxidase with 3% H2O2 for 10 min. The slides were then incubated in blocking solution (serum-free protein block, Dako) for 20 min to block nonspecific binding. The primary antibody was added to the slides at a 1:200 dilution and incubated overnight in a humidified chamber at 40 °C Detection was accomplished using an Envision Horseradish Peroxidase Kit (Dako). Immunostaining was revealed using 3,3′-diaminobenzidine (Dako). The slides were lightly counterstained with haematoxylin, progressively dehydrated through graded alcohols and xylene, and finally covered with a coverslip after mounting in DPX mounting medium. Slides were examined under an Olympus light microscope that was equipped with a ×40 objective.
For mucoricin immunostaining in the lung tissue, heat-induced antigen retrieval was performed to deparaffinize and rehydrate the lung sections through incubation in sodium citrate buffer (10 mM, 0.05% Tween-20, pH 6) for 40 min at 90–95 °C using a steamer (Philips). The tissue sections were allowed to cool for 30 min, washed three times in PBS and permeabilized in 0.2% gelatin/Triton X-100 0.25% for 15 min at room temperature, followed by blocking in 5% BSA/5% normal goat serum (NGS)/PBS for 1 h at room temperature. The tissue slices were subsequently incubated at 4 °C overnight with an anti-mucoricin antibody (2 mg ml−1) in 1% BSA/1% NGS/PBS. The samples were washed three times with PBS then incubated with goat anti-rabbit CF488A or CF555 (1:500, Biotium) in the presence of 200 mg ml−1 CFW for 1 h at room temperature. The tissue sections were incubated for 10 min at room temperature with the nuclear dye TO-PRO-3 (1:2,000, Thermo Fisher Scientific), washed three times with PBS and quenched for autofluorescence by incubation in a solution of 10 mM CuSO4/50 mM NH4Cl.
For mucoricin and CotH3 immunostaining in R. delemar, spores were cultured at 30 °C in RPMI or RPMI supplemented with either 4.5 g dl−1 BSA, 4.5 g dl−1 BSA flow-through or 1 mM caprylic acid for 6-8 h until they swelled. The swollen spores were fixed in 4% paraformaldehyde for 10 min, followed by permeabilization in 0.1% Triton X-100 in PBS for 10 min at room temperature. The permeabilized spores were blocked with 5% NGS in PBS for 1 h at room temperature and subsequently incubated with the anti-mucoricin antibody (10 μg ml−1) or anti-CotH3 antibody (100 μg ml−1)10 for 2 h at room temperature. The spores were washed with Tris-buffered saline (0.01 M Tris HCl/0.15 M NaCl, pH 7.4) containing 0.05% Tween-20 and incubated with goat anti-rabbit CF488A (Biotium) diluted 1:500 in PBS for 1 h at room temperature.
Protein synthesis assessment in R. delemar
R. delemar spores were cultured in black 96-well cell culture plates with a #1.5 glass-like polymer cover slip bottom optimized for high resolution microscopy (P96-1.5P, IBL Baustoff+Labor) at a density of 1.5 × 105 per 200 μl per well. Fungal spores were cultured in RPMI/MOPS (pH 7) supplemented with 0.2% glucose with or without 4.5 g dl−1 BSA flow-through or 2 mM caprylic acid, at 37 °C and 5% CO2. After 2 h (RPMI only) or 3 h (RPMI + BSA flow-through or RPMI + caprylic acid) of culture, media were replenished with OP-puro working solution prepared in the respective media of each treatment group, according to the recommendations of the Protein Synthesis Assay Kit protocol (Cayman Chemical). After 2 h of culture at 37 °C under 5% CO2, R. delemar spores were fixed with the Cell-Based Assay Fixative, washed thrice with the cell-based assay wash buffer and incubated with 5 FAM-azide staining solution in the dark, at room temperature for 30 min. After three washes with cell-based assay wash buffer, R. delemar spores were preserved in 1× assay buffer and imaged directly with a spinning disk confocal microscope (Dragonfly 200, Andor), using the 488 nm/535 nm excitation/emission detection parameters.
Assessment of C12-BODIPY uptake by R. delemar
CFW-labelled R. delemar spores were cultured in RPMI/MOPS (pH 7) for 3 h at 37 °C. To assess whether R. delemar fungus uptakes lipids actively or passively, fungal spores were further cultured at 37 °C or 0 °C for 1 h, in RPMI/MOPS supplemented with 2 μM C12-BODIPY (Invitrogen). Active endocytosis events are inhibited at 0 °C, whereas passive diffusion events remain unaffected80,81. Subsequently, R. delemar spores were washed twice with ice-cold PBS and imaged in PBS using a spinning-disk confocal microscope (Dragonfly 200, Andor). The fluorescence intensity of C12-BODIPY inside R. delemar spores, which correlates with FFA accumulation, was quantified using IMARIS v.10.1 (Oxford Instruments Andor).
Assessment of C11-BODIPY uptake by R. delemar spores
C11-BODIPY (Invitrogen) was pre-oxidized by incubation in 20 mM H2O2/250 μM CuSO4 in UV-grade ethanol at 37 °C under mild shaking for 48 h (ref. 82). As a negative control, an equal amount of C11-BODIPY (40 μM) was incubated in UV-grade ethanol under identical conditions. CFW-labelled R. delemar spores were incubated in RPMI/MOPS (pH 7) without phenol red, supplemented with either pre-oxidized C11-BODIPY or 2 μM control C11-BODIPY, at 37 °C in 18-well μ-Plates (Ibidi). After 4 h, spores were washed twice and imaged in PBS using a spinning-disk confocal microscope (Dragonfly 200, Andor). To validate probe oxidation, spores were first incubated with control C11-BODIPY for 4 h, washed with PBS to remove unbound dye and subsequently treated with increasing concentrations of H2O2 to induce lipid peroxidation. Oxidation was confirmed by a fluorescence shift from 590 nm (red) to 510 nm (green), proportional to the degree of lipid oxidation.
Uptake of oxidized OA by R. delemar spores
For the analysis of OA uptake by R. delemar, fungal spores were cultured in 5% ethanol/RPMI-MOPS supplemented with the indicated concentrations of OA (Sigma-Aldrich) or oxidized OA at 37 °C for 5 h. The spores were collected by scraping, collected in low-affinity 1.5 ml tubes, washed twice with PBS and fixed with methanol-free 4% formaldehyde at room temperature for 20 min. The spores were subsequently washed twice with PBS, stained with 0.5 μg ml−1 Nile red (Thermo Fisher Scientific) at room temperature for 20 min, washed again twice with PBS and stained with 200 μg ml−1 CFW at room temperature for 20 min. Flow cytometry data were acquired using the FACSCanto II system and analysed using FlowJo v.10.6 software (BD Biosciences). All images were acquired using a spinning disk confocal system (Dragonfly 200, Andor) equipped with a motorized inverted Nikon Eclipse Ti2-E microscope, an Andor Sona sCMOS 4.2B-6 camera and four laser lines (405 nm, 488 nm, 561 mm and 633 nm). All images were obtained through z-stacks, deconvolved with Fusion software v.2.3.0.44 (Andor, Oxford Instruments) and further processed in Imaris v.10.1 (Andor, Oxford Instruments) for contrast adjustment, area selection, colour combining and scale bar addition. Certain images were acquired with a Leica TCS SP8 confocal microscope with a ×63 lens and analysed with the use of Fiji/ImageJ. Low-fluorescence immersion oil (Nikon) was used, and imaging was performed at room temperature. Quantitative analysis of fluorescence in confocal microscopy images was performed with IMARIS v.10.1 (Oxford Instruments Andor).
Western blot analysis
The six elutions containing isolated HSA were subjected to western blot analysis for assessment of the isolation protocol. Immunoblotting was performed according to the manufacturer’s instructions using simultaneous antibodies against human transferrin and albumin at a 1:1,000 dilution, followed by incubation at 4 °C overnight. The appropriate secondary antibodies (HRP-conjugated anti-mouse lgG and anti-goat lgG) were used at a 1:5,000 dilution for 1 h at room temperature, and immunoblots were developed by chemiluminescence (ECL; Thermo Scientific). BlueStar Plus Prestained Protein Marker (NIPPON Genetics) was used for the evaluation of protein molecular mass and images were acquired using the ChemiDoc XRSImager (BioRad) system.
Antibodies
Anti-mucoricin and anti-CotH3 antibodies were generated as previously described9. Antibodies against human albumin (sc-365871) and transferrin (sc-365871) were purchased from Santa Cruz. An antibody against mouse active caspase-3 was purchased from Cell Signaling (9661).
Statistical analysis
For univariate and multivariate analysis of the prognostic value of albumin in mucormycosis the primary outcome was all-cause mortality from the time of diagnosis of the infection in the two cohorts at the MDACC, USA and PGIMER, Chandigarh, India and from the time of inclusion to the end of participation of patients (S24 visit) in the Ambizygo study, France. Categorical variables were compared by χ2 or Fisher’s exact test, as appropriate. Continuous variables were compared using the Wilcoxon rank-sum test. Univariate time-dependent survival analysis was performed for the main variables of interest (for example, severe hyperglycaemia, persistent neutropenia, severe hypoalbuminaemia, status of underlying malignancy, steroid use) using Kaplan–Meier curves and the Cox model. The multivariate Cox model was used to identify independent prognostic factor of mortality. Therefore, variables with a P < 0.20 on univariate analysis were included in an initial multivariate model that was then reduced to the final model by backward elimination. Kaplan–Meier survival curves were generated and compared between patients with different albumin levels using the log-rank test. All tests were two-sided with a significance level of 0.05. Data analysis and visualization were performed using Microsoft Office Excel 365 (Microsoft), Prism v10 (GraphPad), SAS v.9.4 (SAS Institute), R (v.4.3.1, R Core Team) and SPSS (IBM, SPSS Statistics for Windows, v.22.0).
For serum lipidomics analysis in Alb−/− and Alb+/+ mice, significant differences between groups were assessed using the Wilcoxon rank-sum test. Data were row-centred and scaled, and visualized as heat maps using the pheatmap R package75. All other statistical analyses were performed using Prism v.10 (GraphPad) unless otherwise stated. P < 0.05 was considered statistically significant for all of the variables tested. A nonlinear regression three-parameter model—log(inhibitor) versus response (three parameters) was used to calculate IC50 values, according to the equation: Y = bottom + (top − bottom)/(\(1+{10}^{(X-\log [{\mathrm{IC}}_{50}])}\)). The FFA concentrations applied to generate the dose–response curves were based on physiological serum concentrations of each FFA in humans24. The statistical methods used to determine significance and the P values of each graph are provided in the figure legends.
Illustrations in Figs. 1f, 2a, 4a,f and 5d, and Extended Data Figs. 1b and 3a were created using BioRender.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.