Animal models
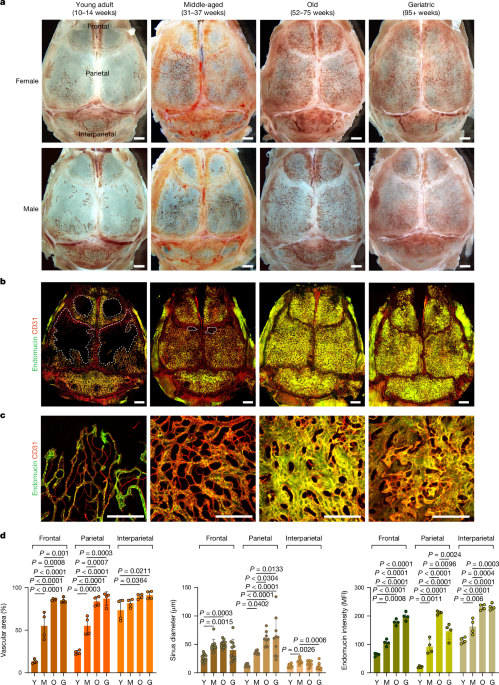
C57BL/6J mice were used for all experiments involving wild-type mice and pharmacological treatments. Mice at the age of 10â14 weeks, 31â37 weeks, 52â75 weeks and >95 weeks were chosen for young adult, middle-aged, old and geriatric groups, respectively. Both female and male mice of all age groups were used for initial BM expansion analyses, while only female mice were used for remaining experiments. Flk1-GFP reporter mice17 were used for initial blood vessel characterization. For genetic labelling of haematopoietic cells, Vav1-cre mice21 were interbred with ROSA26-mTmG reporter mice22 to generate Vav-mTmG mice. For photoconversion of haematopoietic cells, Vav1-cre mice were interbred with ROSA26-CAG-loxP-stop-loxP-KikGR knock-in mice49 to generate Vav-KikGR mice. For pregnancy experiments, 10-week-old C57BL/6J female mice were paired with 10- to 12-week-old C57BL/6J male mice and the onset of pregnancy was determined by the presence of a vaginal plug in the morning. Ten-week-old mice received daily intraperitoneal injections of PTH (1â34) (Bachem, 0.1âmgâkgâ1 for 28 days), PGE2 (Cayman Chemical, 2âmgâkgâ1 for 7 days), AMD3100 (Abcam, 5âmgâkgâ1 for 14 days) before they were euthanized. For DC101 treatment, 10-week-old mice received intraperitoneal injections of DC101 (BioXCell, 40âmgâkgâ1) every 2 days for 12 weeks.
Mice were kept in individually ventilated cages, with constant access to food and water under a 12âh light and 12âh dark cycle regime. Air flow, temperature (21â22â°C) and humidity (55â60%) were controlled by an air management system. Mice were checked daily and maintained in specific pathogen-free conditions. Sufficient nesting material and environmental enrichment was provided. All animal experiments were performed according to the institutional guidelines and laws, approved by local animal ethical committee and were conducted at the Max Planck Institute for Molecular Biomedicine (84-02.04.2016.A160, 81-02.04.2018.A171, 81-02.04.2020.A212, 81-02.04.2020.A416 and 81-02.04.2022.A198), Universitätsmedizin Berlin (G0220/17), Georg-Speyer-Haus (F123/2017) and the University Medical Center Mainz Institute of Transfusion Medicine (G23-1-067 A1TE) under the indicated permissions granted by the Landesamt für Natur, Umwelt und Verbraucherschutz (LANUV) of North Rhine-Westphalia, the State Office for Health and Social Affairs Berlin, Regierungspräsidium Darmstadt and the Landesuntersuchungsamt Rheinland-Pfalz, Germany.
Human subjects, CT acquisition and data analysis
The study was approved by the local ethics committee and the institutional review board (IRB) of Asan Medical Center, and the requirement for informed consent was waived due to the retrospective nature of the study (IRB number: 2023-0658). The study population consisted of 36 patients, divided into 4 groups according to age (between 20 and 40 years, over 60 years) and sex (male, female), with 9 individuals in each group. Patients who underwent CT for evaluation of small cerebral aneurysm from April to May 2023 were eligible. Participants were excluded if they had a previous history of surgery or radiation therapy to the head and neck, vascular or bone-related medical implants, or a suspicious disease other than small cerebral aneurysm.
All human patients underwent CT examinations on the same 128-channel multidetector CT system (Somatom Definition Edge; Siemens). Imaging variables were as follows: 100âkV; 100 effective mAs; axial scan mode; section thickness, 0.5âmm; display FOV, 20.5âcm; pitch, 1; gantry rotation time, 0.5âs; pixel matrix, 512âÃâ512. Images were obtained from the vertex to first cervical spine, without an intravenous injection of contrast media.
The CT data were digitally transferred to a personal computer and processed with ImageJ software (http://rsb.info.nih.gov/ij/). A representative image was selected on a coronal CT image perpendicular to the outermost convex area on an axial CT image. After whole-bone segmentation of the parietal bone, cortical bone and BM were defined by attenuation densities on CT scan: cortical bone as over 850 Hounsfield units and BM as less than 850 Hounsfield units and their areas were calculated.
Sample processing and immunostaining
Mice were euthanized by transcardial perfusion of PBS and 4% paraformaldehyde (PFA), skulls and femur were collected and fixed immediately in ice-cold 4% PFA for 6â8âh under gentle agitation. Bones were decalcified in 0.5âM EDTA for 3 days (for skulls) or 7 days (for femurs) at 4â°C under gentle shaking agitation, washed 5 times in PBS in 5âmin intervals, followed by overnight incubation in cryoprotectant solution (20% sucrose, 2% polyvinylpyrrolidone) and embedding in bone embedding medium (8% gelatin, 20% sucrose, 2% polyvinylpyrrolidone). Samples were stored overnight at â80â°C. 80-μm-thick cryosections were prepared for immunofluorescence staining.
Bone sections were washed in PBS and permeabilized with 0.3% Triton X-100 in PBS for 10âmin at room temperature. Samples were incubated in blocking solution (5% heat-inactivated donkey serum in 0.3% Triton X-100) for 1âh at room temperature. Primary antibodies (rat monoclonal anti-endomucin (V.7C7) (Santa Cruz, sc-65495, 1:200 dilution), rabbit monoclonal anti-vATPaseB1/B2 (Abcam, 200839, 1:200 dilution), goat polyclonal anti-osteopontin (R&D Systems, AF808, 1:200 dilution), goat polyclonal anti-CD31 (R&D, AF3628, 1:200 dilution), rabbit polyclonal anti-caveolin-1 (Cell Signaling, 3238, 1:100), goat polyclonal anti-VEGF164 (R&D Systems, AF-493-NA, 1:200 dilution), and rat monoclonal APC-conjugated anti-CD117 (KIT) (BD Biosciences, 553356, 1:100 dilution) were diluted in PBS with 5% donkey serum and incubated overnight at 4â°C. Next, slides were washed 3â5 times in PBS in 5âmin intervals. Species-specific Alexa Fluor-conjugated secondary antibodies Alexa Fluor 488 (Thermo Fisher Scientific, A21208), Alexa Fluor 594 (Thermo Fisher Scientific, A21209), Alexa Fluor 647 (Thermo Fisher Scientific, A31573 or A21447) diluted 1:500 in PBS with 5% donkey serum were added and incubated overnight at 4â°C. Slides were washed 3â5 times in PBS in 5âmin intervals. Nuclei were counterstained with DAPI (Sigma-Aldrich, D9542, 1:1,000 dilution). Coverslips were mounted with FluoroMount-G (Southern Biotech, 0100-01).
In vivo immunostaining and Evans Blue leakage assay
Rat monoclonal anti-CD31 (BD Biosciences, 553708) was conjugated to Alexa Fluor 647 using the Alexa Fluor 647 Antibody Labeling Kit (Thermo Fisher Scientific, A20186) according to the manufacturerâs instructions. For blood vessel immunostaining, the conjugated anti-CD31 antibody and rat monoclonal PE-conjugated anti-endomucin (V.7C7) (Santa Cruz, 65495 PE) were diluted 1:10 in 200 μl PBS and injected intravenously into the tail vein. For haematopoietic cell immunostaining, rat monoclonal FITC-conjugated anti-CD45 (eBioscience, 11-0451-82), hamster monoclonal FITC-conjugated anti-CD3e (eBioscience, 16-0031-82), rat monoclonal PE-conjugated anti-CD45R/B220 (BD Biosciences, 553090), rat monoclonal FITC-conjugated anti-CD11b (BD Biosciences, 553310) were diluted 1:10 in PBS and injected intravenously into the tail vein. Mice were euthanized 1âh after injection with transcardial perfusion with PBS and 4% PFA and bones were collected and fixed immediately in ice-cold 4% PFA for 6â8âh under gentle agitation. The dura mater was carefully removed from the skull with forceps. Bones were decalcified in 0.5âM EDTA for 1 day (for skulls) or 7 days (for femurs) at 4â°C under gentle shaking agitation, and washed 5 times in PBS in 5âmin intervals. Skulls were counterstained with DAPI (1:500 dilution) for 1âh and trimmed down to the calvarium before mounting with iSpacers (Sunjin Lab, IS011) in PBS. Femurs were cryosectioned, counterstained and mounted as described above.
For the Evans Blue leakage assay, mice were anaesthetized immediately prior to tail vein injection of 200âμl Evans Blue solution (Sigma-Aldrich, E2129, 1% v/w). Mice were euthanized via transcardial perfusion 5âmin after injection as described above. In order to distinguish vascular leakage in the dura mater from the calvarial BM, dura mater tissues were separated from the calvarial bone before overnight decalcification.
Immunostained samples were imaged with a Zeiss LSM980 (Carl Zeiss). Images were analysed, quantified and processed using ZEN Black (Carl Zeiss, v2.3), ImageJ (NIH, v2.0.0) and IMARIS (Bitplane, v10.0.1). Tilescan overview images of skull BM were superimposed on top of a black background, filling empty corners without image data. Vessel diameter was measured by selecting the z-plane image with the widest vessel diameter from the z-stack of the individual vessel.
Scanning electron microscopy
Skull and femur from 12-week-old and 73-week-old mice were isolated and submerged in 4% PFA, 0.5% glutaraldehyde, 2âmM MgCl2, 2âmM CaCl2 in 0.1âM cacodylate buffer, pH 7.4, under agitation for 2âh at room temperature. Samples were fixed further overnight in 2% glutaraldehyde, 2âmM MgCl2, 2âmM CaCl2 in 0.1âM cacodylate buffer, pH 7.4 at 4â°C. Bones were then decalcified over 12 days, changing solution every other day in 5% EDTA in 0.1âM cacodylate buffer, pH 7.4 under rotation at 4â°C. Subsequently, 150âμm sections were generated with a vibratome (VT 1200, Leica). Sections were post-fixed in 1% OsmO4, containing 2.5% PFAâglutaraldehyde mixture buffered with 0.1âM phosphate (pH 7.2) for 5âh and then were placed in graded ethanol for critical-point drying using E3000 (Polaron) critical-point dryer. Critical-point-dried bones were placed on a piece of carbon tape and sputter coated with gold in a SC502 Sputter Coater (Polaron). Specimens were imaged on a Quanta 250 Field Emission Scanning Electron microscope (FEI Quanta 250 FEG, FEI, Hillsboro, OR) installed at the Korea Research Institute of Bioscience and Biotechnology.
Dura mater whole-mount immunostaining
Mice were euthanized by transcardial perfusion of PBS and 4% PFA, skulls were collected and fixed immediately in ice-cold 4% PFA for 6â8âh under gentle agitation. Skulls were decalcified in 0.5âM EDTA for 24âh at 4â°C under gentle shaking agitation, washed 5 times in PBS in 5âmin intervals, trimmed down to the calvarium, and incubated in blocking solution (5% heat-inactivated donkey serum in 0.3% Triton X-100) for 1âh at room temperature. Goat polyclonal anti-CD31 (R&D, AF3628, 1:100 dilution) was diluted in PBS with 5% donkey serum and incubated overnight at 4â°C with gentle agitation. Samples were washed 3â5 times in PBS in 10âmin intervals. Alexa Fluor 647 (Thermo Fisher Scientific, A21447) diluted 1:500 in PBS with 5% donkey serum was added and incubated overnight at 4â°C with gentle agitation. Samples were washed 3â5 times in PBS in 10âmin intervals and mounted with iSpacers (Sunjin Lab, IS011) in PBS.
tMCAO
Sixteen-week-old female C57BL6/J mice were used throughout the experiments. Mice were anaesthetized by intraperitoneal injection of a mixture of 10âmgâkgâ1 xylazine (cp-pharma) and 90âmgâkgâ1 ketamine hydrochloride (cp-pharma). Throughout the whole procedure and during recovery, body temperature was maintained at 37â°C via a heating pad. After ligation of the left proximal common carotid artery and external carotid artery, a 7.0-nylon monofilament (Doccol) with a 0.23-mm coated tip was introduced into the distal internal carotid artery via an incision in the ligated common carotid artery. The monofilament was advanced distal to the carotid bifurcation to occlude the middle cerebral artery. Arter topical application of the local anaesthetic lidocaine hydrochloride (Xylocain Spray 2%, Aspen) the neck wound was closed temporarily for a 45âmin ischaemic period. At reperfusion, the monofilament was withdrawn from the carotid artery and the wound was stitched with 4-0 non-resorbable sutures (Ethibond Excel, Ethicon) and the single s.c. injection of Penicillin G 20 000 U (Benzylpenicillin-Natrium, InfectoPharm) was given. The mouse was returned to its cage to recover under observation.
Chronic myeloid leukaemia
Six-week-old female C57BL/6 mice were purchased from Charles River Laboratories and were used as donors and recipients in all transplants. The transplantation experiments were performed as previously described61. In brief, to induce CML-like myeloproliferative neoplasia, donor BM cells from donor mice pre-treated with 5-fluorouracil (200âmgâkgâ1 intravenously; 4 days prior to collection) were pre-stimulated overnight in medium containing SCF (50ângâmlâ1), IL-6 (10ângâmlâ1) and IL-3 (6ângâmlâ1) and transduced on two consecutive days with murine stem cell virus (MSCV)-IRES-GFP-BCR-ABL1 to induce CML or MSCV-IRES-GFP control virus. Subsequently, transduced cells were intravenously transplanted (2.5âÃâ105 cells per mouse) into sublethally irradiated (900âcGy) recipient mice. Mice were euthanized 14 days after transplantation.
Lineage depletion and transplantation
In order to transplant lineage-negative BM cells, young (10- to 14-week-old) or old (52- to 75-week-old) donor mice were euthanized and skulls were collected. The calvarium was first chopped with scissors in FACS buffer (PBS with 2% fetal calf serum), then crushed with a mortar and pestle. Cell suspension was filtered through a 40-μm mesh filter (Falcon, 352340), resuspended in RBC lysing buffer (Sigma-Aldrich, R7757) for red blood cell lysis and washed with FACS buffer. Cells were resuspended in FACS buffer and incubated with a biotinylated anti-haematopoietic lineage antibody cocktail (Miltenyi-Biotec, 130-092-613, 1:10 dilution), followed by washing with FACS buffer and incubation with R-PE-conjugated streptavidin secondary antibody (Invitrogen, S866, 1:50 dilution). DAPI (1:1,000 dilution) was added to resuspended cells to distinguish live and dead cells and were FACS-sorted for live lineage-negative cells on a FACSAria Fusion (BD Biosciences). Sorted cells were intravenously transplanted (5Ã105 cells/mouse) into lethally irradiated (12âcGy, Best Theratronics, Gammacell 40 Exactor) recipient mice (12-week-old). Mice were euthanized 14 days after transplantation.
FACS analysis of BM and peripheral blood
Mice from each age group were euthanized and skull and femur were collected. Skulls were chopped with scissors in FACS buffer before crushed with mortar and pestle; femurs were crushed without chopping. BM stromal samples were dissociated with Collagenase I (Gibco, 17100-017, 2âmgâmlâ1) and Collagenase IV (Gibco, 17104-019, 2âmgâmlâ1) in PBS for 20âmin at 37â°C with intermittent shaking. Cell suspensions were strained through a 40-μm mesh filter, resuspended in RBC lysing buffer (when applicable) and washed with FACS buffer. Cells were resuspended and incubated with the following primary antibodies in FACS buffer: biotinylated rat monoclonal anti-haematopoietic lineage antibody cocktail (Miltenyi-Biotec, 130-092-613, 1:50 dilution), APC-conjugated rat monoclonal anti-CD117 (BD Biosciences, 553356, 1:100 dilution), FITC-conjugated rat monoclonal anti-CD117 (Biolegend, 105806, 1:100 dilution), FITC-conjugated rat monoclonal anti-Ly6A/E (Sca1) (eBioscience, 11-5981-85, 1:100 dilution), PerCP-Cy5.5-conjugated rat monoclonal anti-Ly-6A/E (Invitrogen, 45-5981, 1:100), APC-Cy7-conjugated hamster monoclonal anti-CD48 (BD Biosciences, 561242, 1:100 dilution), PE-conjugated rat monoclonal anti-CD150 (SLAM) (Biolegend, 115904, 1:100 dilution), Alexa Fluor 647-conjugated rat monoclonal anti-CD150 (Biolegend, 115918, 1:100 dilution), PE-Cy7-conjugated rat monoclonal anti-CD45 (eBioscience, 25-0451-82, 1:100 dilution), BV421-conjugated rat monoclonal anti-TER-119 (Biolegend, 116234, 1:100 dilution), FITC-conjugated rat monoclonal anti-CD71 (Biolegend, 113806, 1:100 dilution), Alexa Fluor 647-conjugated rat monoclonal anti-CD31 (BD Biosciences, 553708, conjugation described above, 1:200 dilution), PE-conjugated rat monoclonal anti-endomucin (Santa Cruz, 65495 PE, 1:100 dilution), PE-Cy7-conjugated rat monoclonal anti-CD16/32 (eBioscience, 25-0161, 1:100 dilution), eFluor 450-conjugated rat monoclonal anti-CD34 (eBioscience, 48-0341, 1:100 dilution), PE-conjugated rat monoclonal anti-CD127 (eBioscience, 12-1271, 1:100 dilution), BV711-conjugated rat monoclonal anti-CD41 (BD Biosciences, 740712, 1:100 dilution), PE-conjugated rat monoclonal anti-CD105 (eBioscience, 12-1051-82, 1:100 dilution), APC-conjugated hamster monoclonal anti-CD3e (eBioscience, 17-0031, 1:100 dilution), PE-conjugated rat monoclonal anti-CD45R/B220 (BD Biosciences, 553090, 1:100 dilution), FITC-conjugated rat monoclonal anti-CD11b (BD Biosciences, 553310, 1:100 dilution). Cells were washed, resuspended in FACS buffer with Alexa Fluor 405-conjugated (Invitrogen, S32351, 1:100 dilution) or APC-Cy7-conjugated (BD Biosciences, 554063, 1:100 dilution) streptavidin secondary antibody, washed again before analysis with a FACSymphony A5 Cell Analyzer (BD Biosciences).
Peripheral blood was collected from the submandibular vein with lancets (Medipoint) into EDTA-coated tubes. Blood was resuspended in RBC lysing buffer and washed with FACS buffer. Cells were resuspended and incubated with the following primary antibodies in FACS buffer: biotinylated rat monoclonal anti-haematopoietic lineage antibody cocktail (Miltenyi-Biotgec, 130-092-613, 1:50 dilution), APC-conjugated rat monoclonal anti-CD117 (BD Biosciences, 553356, 1:100 dilution), FITC-conjugated rat monoclonal anti-Ly6A/E (Sca1) (eBioscience, 11-5981-85, 1:100 dilution), Pacific Blue-conjugated mouse monoclonal anti-CD45.2 (Biolegend, 109820, 1:100 dilution), APC-conjugated hamster monoclonal anti-CD3e (eBioscience, 17-0031, 1:100 dilution), PE-conjugated rat monoclonal anti-CD45R/B220 (BD Biosciences, 553090, 1:100 dilution), FITC-conjugated rat monoclonal anti-CD11b (BD Biosciences, 553310, 1:100 dilution). Cells were washed and resuspended in FACS buffer before analysis with a FACSymphony A5 Cell Analyzer (BD Biosciences).
RNA extraction and quantitative PCR
FACS-sorted cells from 10-week-old mouse skulls were lysed and RNA was extracted using a Monarch Total RNA Miniprep Kit (New England BioLabs, T2010S). Extracted RNA concentration was measured with a NanoDrop 8000 Spectrophotometer (Thermo Fisher Scientific) and cDNA was generated with a LuncaScript RT SuperMix Kit (New England BioLabs, E3010L). Quantitative PCR with reverse transcription was performed with a BioRad CFX96 real-time PCR system using FAM-conjugated Taqman probes for Vegfa (Mm00437306_m1) or using PowerUp SYBR Green Master Mix (Applied Biosystems, A25742) with primers designed using Pimer-BLAST or adopted from previously published studies: Vegfa120 (5â²-AACGATGAAGCCCTGGAGTG-3â²; 5â²-TGAGAGGTCTGGTTCCCGA-3â²); Vegfa164 (5â²-AACGATGAAGCCCTGGAGTG; 5â²-GACAAACAAATGCTTTCTCCG-3â²); Vegfa188 (5â²-AACGATGAAGCCCTGGAGTG-3â²; 5â²-AACAAGGCTCACAGTGAACG-3â²). Gene expression levels were normalized to the endogenous VIC-conjugated Gapdh probe (44326317E) as control.
ELISA
Mice from each age group were euthanized, bones were collected. Skulls were chopped before being crushed with a mortar and pestle in ice-cold RIPA lysis buffer; femurs were crushed without chopping. Supernatants of centrifuged lysates were further concentrated using an Ultra-0.5 Centrifugal Filter Unit with a 3âkDa cutoff (Millipore, UFC500396), resulting concentrations were measured using a Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, 23225), and the concentrations of VEGFA in tissue extracts were measured using a Mouse VEGFA Quantikine ELISA Kit (R&D Systems, MMV00-1).
Hypoxia analysis
Hypoxic cells were detected with the hypoxia probe pimonidazole (Pimo, Hypoxyprobe) according to the manufacturerâs instructions. Mice were intraperitoneally injected with 60âmgâkgâ1 1âh before analysis.
VEGFA plasmid construction and overexpression
To generate the pLIVE-VEGFA165-HA-MP-Asp8x bone-homing protein containing VEGF165 fused to a HA tag, metalloprotease and 8x Asp peptide sequences, a cDNA fragment encoding amino acids 1â191 of human VEGFA was amplified via PCR using the following oligonucleotide primers: VEGFA-AscI-Fwd: 5â²-ATGAACTTTCTGCTGTCT-3â² and VEGFA-XhoI-Rev: 5â²-CCGCCTCGGCTTGTCACATCTGCA-3â² and annealed with the NEBuilder Assembly Cloning Kit.
Ten-week-old mice were used for hydrodynamic tail vein injection. Mice were injected with 0.5âμgâgâ1 (plasmid/body weight) pLIVE-Vegfa plasmid suspended in TransIT-EE hydrodynamic delivery solution (Mirus, MIR5340). The appropriate amount of plasmid was suspended in an injection volume of 10% of the body weight and injected into each individual mouse via the tail vein in 5â7âs as previously reported62.
Adipocyte analysis
To stain for neutral lipids, the entire calvarium or femur cryosections were incubated in BODIPY 493/503 (Invitrogen, D3922; 1:1,000 dilution) for 1âh at room temperature with gentle agitation (only calvarium). Samples were washed with PBS 3â5 times at 5âmin intervals before mounting.
Analysis of inflammatory cytokines
Mice from each age group were euthanized and bones were collected. Skulls were chopped before being crushed with a mortar and pestle in ice-cold RIPA lysis buffer; femurs were crushed without chopping. Supernatants of centrifuged lysates were further concentrated using an Ultra-0.5 Centrifugal Filter Unit with a 3âkDa cutoff (Millipore, UFC500396), resulting concentrations were measured using a Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, 23225), and concentrations of inflammatory cyotokines were measured with LEGENDplex Mouse Inflammation Panel (13-plex) with V-bottom plates (Biolegend, 740446). Analysis on a FACSymphony (BD Biosciences) and quantification were performed according to the manufacturerâs protocol. Data analysis was performed using software provided by Biolegend. Manual gating was used to define beads A and B, and automatic gating strategy was used to gate individual cytokines in the APCâPE plot.
Irradiation with partial shielding
Mice were anaesthetized with ketamine (100âmgâkgâ1) and xylazine (10âmgâkgâ1) prior to irradiation. For partial shielding, the entire head or both legs of a mouse were inserted into the opening of the cylindrical 1-inch-thick lead shield (JRT Associates, PTI-50-P) and exposed to lethal irradiation (12âcGy). The mouse was returned to its cage to recover under observation.
Sample preparation for scRNA-seq
Mice from each age group were euthanized and skull and femur were collected. Skulls were chopped with scissors in FACS buffer before crushed with mortar and pestle; femurs were crushed without chopping. BM stromal samples were dissociated with Collagenase I (Gibco, 17100-017, 2âmgâmlâ1) and Collagenase IV (Gibco, 17104-019, 2âmgâmlâ1) in PBS for 20âmin at 37â°C with intermittent shaking. Cell suspensions were strained through a 40-μm mesh filter and washed with FACS buffer. Cells were resuspended and incubated with biotinylated rat monoclonal anti-haematopoietic lineage antibody cocktail (Miltenyi-Biotec, 130-092-613, 1:50 dilution). Cells were washed, resuspended in FACS buffer with mouse monoclonal anti-Biotin MicroBeads (Miltenyi-Biotec, 130-105-637, 1:50 dilution) and incubated before being loaded into a magnetic-associated cell sorting (MACS) column (Miltenyi-Biotec, 130-042-201) for lineage depletion. Linâ cells were further incubated with rat monoclonal anti-CD45 MicroBeads (Miltenyi-Biotec, 130-052-301, 1:50 dilution), rat monoclonal anti-CD117 MicroBeads (Miltenyi-Biotec, 130-091-224, 1:50 dilution), biotinylated rat monoclonal anti-CD71 (Biolegend, 113803, 1:100). Cells were washed, resuspended in FACS buffer with mouse monoclonal anti-Biotin MicroBeads (Miltenyi-Biotec, 130-105-637, 1:50 dilution) and incubated before being loaded into a magnetic-associated cell sorting (MACS) column (Miltenyi-Biotec, 130-042-201) for further haematopoietic depletion. Single-cell suspensions were processed with BD Rhapsody and scRNA-seq libraries were evaluated and quantified by Agilent Bioanalyzer using High Sensitivity DNA Kit (Agilent Technologies, 5067-4626) and Qubit (Thermo Fisher Scientific, Q32851). Individual libraries were diluted to 4ânM and pooled for sequencing. Pooled libraries were sequenced by using High Output Kit (Illumina, TG-160-2002) with a NextSeq500 sequencer (Illumina).
scRNA-seq
Preprocessing: STAR version 2.7.10a (PMID: 23104886) was used to generate a reference genome index for GRCm39, with Gencode annotations vM29, subset to lncRNA and protein-coding genes.
FASTQ reads were mapped against the reference genome index using STAR with the settings â–soloType CB_UMI_Complex –soloCellFilter None –outSAMtype BAM SortedByCoordinate –soloFeatures GeneFull_Ex50pAS –soloCBmatchWLtype 1MM –soloUMIlen 8 –soloCBwhitelist BD_CLS1.txt BD_CLS2.txt BD_CLS3.txt –runRNGseed 1 –soloMultiMappers EM –readFilesCommand zcat –outSAMattributes NH HI AS nM NM MD jM jI MC ch CB UB GX GN sS CR CY UR UYâ. Libraries using standard BD Rhapsody beads were mapped using the adapter parameters â–soloAdapterSequence NNNNNNNNNACTGGCCTGCGANNNNNNNNNGGTAGCGGTGACA –soloCBposition 2_0_2_8 2_21_2_29 3_1_3_9 –soloUMIposition 3_10_3_17â, libraries with BD Rhapsody enhanced beads with –soloAdapterSequence NNNNNNNNNGTGANNNNNNNNNGACA –soloCBposition 2_0_2_8 2_13_2_21 3_1_3_9 –soloUMIposition 3_10_3_17.
Raw counts were imported as AnnData63 objects. We removed low complexity barcodes with the knee plot method, and further filtered out cells with a mitochondrial mRNA content, as well as unusually high total and gene counts using manually determined cutoffs for each sample. Doublets were scored with scrublet64. Finally, each sampleâs gene expression matrix was normalized using scran65 (1.22.1) with Leiden clustering66 input at resolution 0.5.
G2M and S phase scores were assigned to each cell using gene lists from ref. 67 and the scanpy68 (1.9.6) sc.tl.score_genes_cell_cycle function.
Embedding, clustering and annotation: different combinations of samples and cell populations (all, ECs, HSCs), were used as input for 2D embedding and clustering: the corresponding expression matrix was subset to the 2,000 most highly variable genes (sc.pp.highly_variable_genes, flavour âseuratâ). The top 50 principal components were calculated, and batch-corrected using Harmony69 (0.0.9). The principal components served as basis for k-nearest neighbour calculation (sc.pp.neighbors, n_neighbors=30), which were used as input for UMAP70 layout (sc.tl.umap, min_dist=0.3). Cell populations were clustered using scanpy.tl.leiden, and a suitable resolution was chosen for a first-pass annotation. Here, contaminating cell populations, including multiplet clusters, were removed, and clustering was repeated. Cluster marker genes were calculated using a pseudobulk approach, comparing aggregate counts with 2 pseudoreplicates for each cluster to all remaining cells (pyDeSEQ2 0.4.8). Finally, expression of select marker genes was plotted using Matplotlib71 (3.8.4) imshow, and clusters were annotated accordingly.
Differential expression analysis: Differentially expressed genes were calculated using a pseudobulk approach, comparing aggregate counts with two pseudoreplicates for each condition (pyDeSEQ2 0.4.8).
Skull BM photoconversion
Vav1-KikGR mice were anaesthetized with ketamine (100âmgâkgâ1) and xylazine (10âmgâkgâ1). A skin flap was generated to expose the calvarium, as previously described72. Each exposed area of the calvarium was then exposed to UV light from a Zeiss Axio Imager (Zeiss Microscopy) for 60âs, confirmed for photoconversion from green to red fluorescence, before exposing another area. The skin flap was sutured back together and peripheral blood was analysed by flow cytometry, as described above, to check for the presence of photoconverted cells, which were non-existent in the peripheral blood immediately after photoconversion. One week after photoconversion, peripheral blood was drawn, stained for Alexa Fluor-conjugated rat monoclonal anti-CD45, and was analysed by flow cytometry for CD45+ photoconverted haematopoietic cells derived from the skull BM.
Statistical analysis
No statistical methods were used to predetermine sample size. The experiments were randomized and investigators were blinded to allocation during experiments and outcome analyses. All values are presented as meanâ±âs.d. Statistical significance was determined by the two-tailed unpaired Studentâs t-test between two groups or the Tukey multiple comparison test (one-way ANOVA) for multiple-group comparison. Statistical analyses were performed using GraphPad Prism 9.0 (GraphPad Software). Statistical significance was set at Pâ<â0.05.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.