Data reporting
No statistical methods were used to predetermine sample size. The experiments were not randomized, and the investigators were not blinded to allocation during experiments and outcome assessment.
Clinical sample acquisition
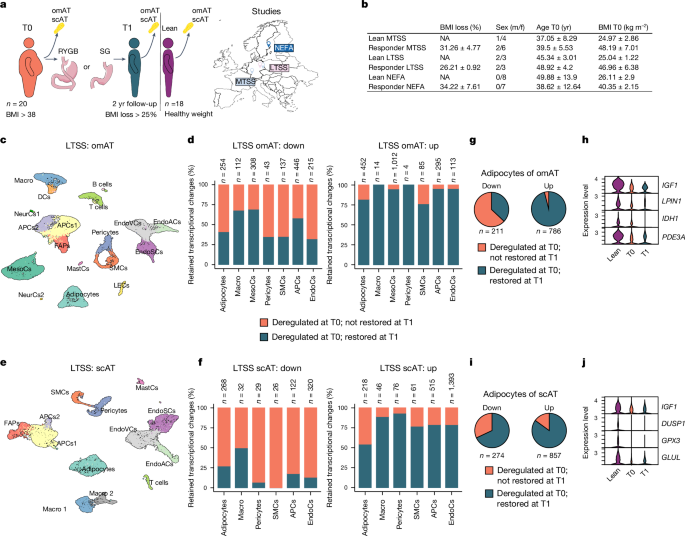
Human AT biopsies were obtained from three independent studies: MTSS, LTSS and NEFA.
MTSS
The MTSS samples comprised samples from omental visceral AT biopsies obtained in the context of a two-step BaS treatment, which included a sleeve gastrectomy as the first step (T0) and laparoscopic RYGB as the second step (T1)16. Individuals with syndromal, monogenic, early-onset obesity or individuals with other known concurrent diseases, including acute infections or malignant diseases, were not included in the study. Individuals were not required to adhere to any specific diet before or after surgery but received individual dietary recommendations during regular visits in the obesity management centre. Insulin resistance was determined using a hyperinsulinaemicâeuglycaemic clamp technique or the homeostatic model assessment for insulin resistance (HOMA-IR). Only biopsies from individuals that (1) lost 25% or more of BMI between T0 and T1 (Extended Data Table 1), (2) had undergone surgery at the Municipal Hospital Karlsruhe or Municipal Hospital Dresden-Neustadt, (3) were not diagnosed with diabetes, and (4) did not receive any glucose-lowering medication were used for snRNA-seq in this study. AT samples were collected during elective laparoscopic abdominal surgery as previously described63, snap-frozen in liquid nitrogen and stored at â80â°C. Body composition and metabolic parameters were measured as previously described64. Samples of healthy individuals who were not obese were collected during routine elective surgeries such as herniotomies, explorative laparoscopies and cholecystectomies at the same hospitals. The study was approved by the Ethics Committee of the University of Leipzig under approval number 159-12â21052012 and was performed in agreement with the Declaration of Helsinki.
LTSS
The human study samples comprised samples from omental visceral and subcutaneous abdominal AT, collected in the context of a two-step BaS treatment. Following an initial sleeve gastrectomy (T0), a laparoscopic RYGB was made in the second step (T1)16. Individuals with syndromal, early-onset obesity or individuals with other known concurrent diseases, including acute infections or malignant diseases, were not included in the study. Individuals did not adhere to any specific diet before or after surgery but received individual healthy diet recommendations during regular visits in the obesity management centre. Insulin resistance was determined using HOMA-IR. Only individuals that (1) lost 25% or more of BMI between T0 and T1 (Extended Data Table 1), (2) had undergone surgery at the Leipzig University Hospital, (3) were not diagnosed with diabetes and (4) did not receive any glucose-lowering medication were included. AT samples were collected during elective laparoscopic abdominal surgery as previously described63, snap-frozen in liquid nitrogen and stored at â80â°C. Body composition and metabolic parameters were measured as previously described64. Samples from healthy donors that were not obese were collected during routine elective surgeries (herniotomies, explorative laparoscopies, cholecystectomies) at the same hospital. The study was approved by the Ethics Committee of the University of Leipzig under approval number 159-12â21052012 and performed in agreement with the Declaration of Helsinki.
NEFA study
The NEFA study (NCT01727245) comprises samples from subcutaneous abdominal AT from individuals before and after RYGB surgery, as well as healthy controls who had never been obese8,65. For this, biopsies were obtained under local anaesthesia before (T0) and 2âyr post-surgery (T1). Only samples from individuals that (1) lost more than 25% BMI between T0 and T1, (2) were not diagnosed with diabetes at T0 and T1 and (3) did not take glucose-lowering medication were included in the present study (Extended Data Table 1). Samples from control subjects were obtained from individuals that were BMI- and age-matched to RYGB patients at T1 as reported previously8. AT samples were handled as reported before65, snap-frozen in liquid nitrogen and stored at â80â°C. The study was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committee of the Karolinska Institute, Stockholm (approval number 2011/1002-31/1).
Mice
All mice were kept on a 12-h/12-h light/dark cycle at 20â60% (23â°C) humidity in individually ventilated cages, in groups of between two and five mice, in a pathogen-free animal facility in the SLA building at ETH Zurich. The health of mice was monitored closely, and any mouse exhibiting persistent clinical signs of ill health or distress was excluded from this study. The 16- and 29-week-old male C57BL/6J diet-induced obesity mice (catalogue no. 380050) and diet-induced obesity control mice (catalogue no. 380056) were obtained from The Jackson Laboratory and were kept on the respective diets for another 2âweeks until tissue harvest or diet switch. Different mice were used for insulin tolerance tests and glucose tolerance tests. AdipoERCre66 and NuTRAP67 mice were maintained on a C57BL/N background. Homozygous NuTRAP and AdipoERCre mice were bred to generate AdipoERCre x NuTRAP mice. AdipoERCre x NuTRAP mice were kept on HFD or chow diet for 12 or 25âweeks before tissue harvest or diet switch. The HFD used contained 60% (kcal%) fat (diet no. 2127, Provimi Kliba); the low-fat chow diet used contained 10% (kcal%) fat (diet no. 2125, Provimi Kliba). During the WL period both experimental groups received chow diet (diet no. 3437, Provimi Kliba). All animal experiments were approved by the Cantonal Veterinary Office, Zurich.
Tamoxifen application
The 4â5-week-old AdipoERCre x NuTRAP mice were gavaged two times with 1âmg of tamoxifen dissolved in corn oil. Tamoxifen was washed out for 2âweeks before starting HFD.
Physiological measurements
Glucose tolerance test
Mice were fasted for 6âh during dark phase before administration of 1âg of glucose per kg body weight by intraperitoneal injection. Blood was collected from the tail vein at 0, 15, 30, 60, 90 and 120âmin and blood glucose concentrations were measured using an Accu-Check Aviva glucometer.
Insulin tolerance test
Mice were fasted for 6âh during dark phase before administration of 1âU per kg body weight of human insulin (insulin Actrapid HM, Novo Nordisk) by intraperitoneal injection. Blood was collected from the tail vein at 0, 15, 30, 60, 90 and 120âmin and blood glucose concentrations were measured using a Accu-Check Aviva glucometer.
In vivo indirect calorimetry
Measurements were obtained from one 8-cage and one 16-cage Promethion Core Behavioral System that were in the same room. Mice were habituated to the system for 36âh before measurements were started.
Live body composition
Mice were fasted for 6âh during dark phase. Live mouse body composition was measured with a magnetic resonance imaging technique (EchoMRI 130, Echo Medical Systems). Fat and lean mass were analysed using EchoMRI 14 software.
Fasting insulin
EDTA plasma was isolated from fasted blood samples (fasting 6âh). Insulin was measured with Ultra Sensitive Mouse Insulin ELISA Kit (Crystal Chem, catalogue no. 90080).
Postprandial insulin
EDTA plasma (50âµl) was thawed on ice and used in a custom U-PLEX assay (Meso Scale Discovery) according to the manufacturerâs instructions. A Mesoscale SI 2400 was used to read the plate.
Postprandial leptin
EDTA plasma (50âµl) was thawed on ice and used in a custom U-PLEX assay (Meso Scale Discovery) according to the manufacturerâs instructions. A Mesoscale SI 2400 was used to read the plate.
Liver triglycerides
First, 50âmg of frozen liver was homogenized in 1âml of isopropanol, lysed for 1âh at 4â°C and centrifuged for 10âmin at 2,000g at 4â°C. The supernatant was transferred into a new tube and stored at â80â°C until use. Triglyceride levels were measured by mixing 200âµl of reagent R (Monlab, catalogue no. SR-41031) and 5âµl of sample or Cfas calibrator dilutions (Roche, catalogue no. 10759350; lot no. 41009301), then incubating for 10âmin while shaking at room temperature and measuring optical density at 505 nm (OD505) with a plate reader (BioTek Gen5 Microplate Reader).
Cell culture experiments
AT digestion
AT was minced and digested at 37â°C while shaking in collagenase buffer (25âmM NaHCO3, 12âmM KH2PO4, 1.3âmM MgSO4, 4.8âmM KCl, 120âmM NaCl, 1.2âmM CaCl2, 5âmM glucose, 2.5% BSA; pH 7.4) using 2âmg of collagenase type II (Sigma-Aldrich, catalogue no. C6885-1G) per 0.25âg of tissue. After 30âmin tissues were resuspended, and for ingAT digestion continued for 15âmin whereas epiAT was processed immediately. An equal volume of growth medium (DMEM (Gibco, catalogue no. 31966021), 10% FBS (Gibco, catalogue no. 10500-064, Lot no. 2378399H), 1% penicillin-streptomycin (Gibco, catalogue no. 15140-122)) was added and digested tissue was centrifuged for 4âmin at 300g, and the floating fraction was transferred into a new Falcon tube and kept at 37â°C. The SVF was resuspended in 5âml of erythrocyte lysis buffer (154âmM NH4Cl, 10âmM NaHCO3, 0.1âmM EDTA, 1% penicillin-streptomycin), incubated at room temperature for 5âmin, filtered through a 40âµM mesh filter and centrifuged for 5âmin, 300g. The SVF was resuspended in growth medium and counted.
SVF differentiation
A total of 10,000 cells were plated into one well of a collagen-coated (Sigma-Aldrich, catalogue no. C3867) 96-well plate and kept in culture until they reached confluency, with media change every 48âh. At 2âd post-confluence, medium was changed to induction medium (DMEM, 10% FBS, 1% penicillin-streptomycin, 10ânM insulin (Sigma-Aldrich, catalogue no. I9278), 0.5âmM 3-isobutyl-1-methylxanthin (Sigma-Aldrich, catalogue no. I7018-1G), 1âµM dexamethasone (Sigma-Aldrich, catalogue no. D4902), 1âµM rosiglitazone (Adipogen, catalogue no. AG-CR1-3570-M010)). After 48âh medium was changed to maintenance medium (DMEM, 10% FBS, 1% penicillin-streptomycin, 10ânM insulin). Medium was changed every 48âh for 8âd.
AdipoRed assay
The SVF was cultured as described and controls were either kept in growth medium or only maintenance medium without induction. On day 8 after induction, cells were washed twice in PBS, and AdipoRed (Lonza, catalogue no. LZ-PT-7009) reagent was used according to the manufacturerâs instructions and read with a plate reader (BioTek Gen5 Microplate Reader).
Primary adipocyte culture
Primary floating adipocytes were cultured under membranes according to Harms et al.68. Packed adipocytes (30âµl) were seeded onto one membrane and kept in inverted culture for 48âh in maintenance medium (DMEM-F12 (Gibco, catalogue no. 31330095), 10% FBS, 1% penicillin-streptomycin, 10ânM insulin). After 48âh of maintenance, adipocytes were washed and serum and glucose starved overnight in KREBBS-Ringer buffer (120âmM NaCl, 4.7âmM KCl, 1.2âmM KH2PO4, 1.2âmM MgSO4, 2.5âmM CaCl2, 25âmM HEPES (Lonza, catalogue no. BEBP17-737E), pH 7.4) and 2.5% fat-free BSA (Sigma-Aldrich, catalogue no. A6003).
Glucose uptake
Glucose uptake from primary adipocytes was measured using the Glucose Uptake-Glo Assay Kit (Promega, catalogue no. J1341) according to the manufacturerâs instructions. Adipocytes were preincubated with 5ânM insulin for 15âmin before 2-deoxy-d-glucose was added at 1âmM final concentration. Protein concentration was measured using a Pierce 660ânm Protein Assay Kit (Thermo Fisher, catalogue no. 22662) and the Ionic Detergent Compatibility Reagent (Thermo Fisher, catalogue no. 22663). Both assays were read with a plate reader (BioTek Gen5 Microplate Reader).
C16 uptake
Starved adipocytes were incubated with 5ânM BODIPY-palmitate (Thermo Fisher, catalogue no. D3821) in the presence of 10ânM insulin for 1âh. Subsequently, adipocytes were washed twice and lysed in 200âµl of RIPA buffer. Then, 100âµl of lysate was used to measure BODIPY signal. Diluted lysate was used to measure protein concentration using a DC Protein Assay Kit II (Bio-Rad Laboratories, catalogue no. 5000112) for normalization. Both assays were read with a plate reader (BioTek Gen5 Microplate Reader).
Histology
Tissues were collected, fixed in 4% PBS-buffered formalin for 72âh at 4â°C and stored in PBS at 4â°C. Following paraffin embedding, tissues were sent to the pathology service centre at Instituto Murciano de Investigación Biosanitaria Virgen de la Arrixaca for sectioning, trichrome staining, haematoxylin and eosin staining, and imaging. Tissues from two independent experiments were sent for sectioning.
Adipocyte size quantification
Images of ingAT and epiAT were taken with 3DHISTECH Slide Viewer 2 and then analysed with Adiposoft69 using Fiji ImageJ70. Five to ten images were taken of each section belonging to a biological replicate (nâ=â4).
Sample processing and library preparation
Isolation of nuclei from mouse tissue
Nuclei were isolated from snap-frozen epiAT in ice-cold Nuclei Extraction Buffer (Miltenyi, catalogue no. 130-128-024) supplemented with 0.2âUâµlâ1 recombinant RNase Inhibitor (Takara, catalogue no. 2313) and 1à cOmplete EDTA-free Protease Inhibitor (Roche, catalogue no. 5056489001) using the gentleMACS Octo Dissociator (Miltenyi, catalogue no. 130-096-427), using C-tubes (Miltenyi, catalogue no. 130-093-237). Nuclei were subsequently filtered through a 50âµm cell strainer (Sysmex, catalogue no. 04-0042-2317) and washed two times in PBS-BSA (1% w/v) containing 0.2âUâµlâ1 RNase inhibitor. For snRNA-seq, five mice were pooled per condition.
Isolation of nuclei from human tissue
Nuclei were isolated from snap-frozen human AT (10â50âmg) in ice-cold Nuclei Extraction Buffer (Miltenyi, catalogue no. 130-128-024) supplemented with 1âUâµlâ1 recombinant RNase Inhibitor (Takara, catalogue no. 2313), 1à cOmplete EDTA-free Protease Inhibitor (Roche, catalogue no. 5056489001) and 10âmM sodium butyrate using the gentleMACS Octo Dissociator (Miltenyi, catalogue no. 130-096-427), using C-tubes (Miltenyi, catalogue no. 130-093-237).
The nuclei suspension was filtered through a 50âµm strainer, supplemented with PBS-BSA (1% w/v) containing 1à protease inhibitor and RNase inhibitor and centrifuged at 4â°C, at 500g for 10âmin. The nuclei pellet was resuspended in 1âml of PBS-BSA (1%, w/v) supplemented with RNase inhibitor (0.5âUâµlâ1) and 1à protease inhibitor and was transferred into a new 1.5âml tube.
snRNA-seq of AT
Nuclei were counted using a haemocytometer and Trypan blue, concentration was adjusted to approximately 1,000 nuclei per µl and they were loaded onto a G-chip (10x Genomics, catalogue no. PN-1000127). Single-cell gene expression libraries were prepared using the Chromium Next GEM Single Cell 3â² v3.1 kit (10x Genomics) according to the manufacturerâs instructions. To accommodate for low RNA content, two cycles were added to the complementary DNA amplification PCR. Libraries were pooled equimolecularly and sequenced in PE150 (paired-end 150) mode on a NovaSeq 6000 with about 40,000 reads per nucleus at Novogene or using a NovaSeqX at the Functional Genomics Center, Zurich.
Paired TRAPâseq, CUT&Tag and ATACâseq
Paired TRAPâseq, CUT&Tag and ATACâseq protocols were developed on the basis of published protocols67,71,72,73,74.
Ribosome and nuclei isolation
Nuclei and ribosomes were isolated from snap-frozen epiAT from AdipoERCre x NuTRAP mice in ice-cold Nuclei Extraction Buffer (Miltenyi, catalogue no. 130-128-024) supplemented with 0.2âUâµlâ1 recombinant RNase Inhibitor (Takara, catalogue no. 2313), 1à cOmplete EDTA-free Protease Inhibitor (Roche, catalogue no. 5056489001) and 10âmM sodium butyrate using the gentleMACS Octo Dissociator (Miltenyi, catalogue no. 130-096-427), using C-tubes (Miltenyi, catalogue no. 130-093-237). The nuclei suspension was filtered through a 50âµm strainer and centrifuged at 4â°C, 500g for 5âmin. The supernatant was transferred into a new tube and supplemented with 2âmM dithiothreitol, 100âµgâmlâ1 cycloheximide (Sigma-Aldrich, catalogue no. 01810) and 1âmgâmlâ1 sodium heparin (Sigma-Aldrich, catalogue no. H3149-10KU) and kept on ice. The nuclei pellet was resuspended in 1âml of PBS-BSA (1%, w/v) supplemented with 0.2âUâµlâ1 RNase inhibitor, 1à cOmplete EDTA-free Protease Inhibitor and 10âmM sodium butyrate and transferred into a new 1.5âml tube. Nuclei were centrifuged and subsequently bound to Dynabeads MyOne Streptavidin C1 beads (Thermo Fisher, catalogue no. 65002) for 30âmin at 4â°C followed by three washes with PBS-BSA (1% w/v).
TRAPâseq
Per sample, 25âµl of GFP-Trap Magnetic Agarose Beads (ChromoTEK, catalogue no. gtma-20) were washed in 2âml of polysome lysis buffer (50âmM TRIS-HCl pH 7.5, 100âmM NaCl, 12âmM MgCl2, 1% Igepal CA-630 (Sigma-Aldrich, catalogue no. I8896), 1à protease inhibitor). The supernatant was mixed with the beads and incubated at 4â°C on a rotator for 1â2âh. Subsequently, tubes were put on a magnetic stand and the supernatant was removed. The beads were washed three times with polysome lysis buffer supplemented with 2âmM dithiothreitol (Sigma-Aldrich, catalogue no. D0632-10G), 100âµgâmlâ1 cycloheximide (Sigma, catalogue no. D0632-10G) and 1âmgâmlâ1 sodium heparin (VWR, catalogue no. ACRO411210010) and resuspended in 1âml Trizol (Thermo Fisher, catalogue no. 15596). Trizol preserved samples were kept at â80â°C until RNA isolation. RNA was isolated by adding 200âµl of chloroform (Sigma-Aldrich, catalogue no. 288306) to samples, followed by shaking and centrifugation at 4â°C, 12,000g for 15âmin. The aqueous phase was transferred into a new tube and RNA was isolated and DNase treated with the RNA Clean and Concentrator-5 kit (Zymo Research, catalogue no. R1016), following the manufacturerâs instructions.
RNA libraries were prepared by performing reverse transcription and template switching using Maxima H Minus reverse transcriptase (Thermo Fisher, catalogue no. EP0753), a template switch oligo and an oligodT primer to generate full-length cDNA. cDNA was amplified using the KAPA Hotstart 2x ReadyMix (Roche Diagnostics, catalogue no. 7958935001). Then, 1â3âng of cDNA was tagmentated using 1.3âµg of Tn5 and amplified using KAPA HiFi plus dNTPs (Roche Diagnostics, catalogue no. 07958846001) and the following PCR settings: 72â°C 5âmin, 98â°C 30âs, 10 cycles of 98â°C for 10âs, 63â°C for 30âs, 72â°C for 1âmin, hold at 4â°C. Libraries were quantified using the KAPA library quantification kit (Roche Diagnostics, catalogue no. 079602), and sequenced in PE150 mode on a NovaSeq 6000 at Novogene.
CUT&Tag
CUT&Tag was performed as previously described with minor adjustments74,75. All buffers were supplemented with 1 x cOmplete EDTA-free Protease Inhibitor and 10âmM sodium butyrate. Briefly, nuclei bound to beads were aliquoted into 96-well LoBind plates (Eppendorf, catalogue no. 0030129547) and incubated with primary antibodiesâanti-H3K4me3 (abcam, catalogue no. ab8580), anti-H3K27me3 (Cell Signaling Technology, catalogue no. C36B11), anti-H3K27ac (abcam, catalogue no. ab4729), anti-H3K4me1 (abcam, catalogue no. ab8895)âovernight at 4â°C. With the plate on a magnet, the primary antibody solution was removed, and the beads were resuspended in secondary antibody solution (guinea pig anti-rabbit IgG (antibodies-online, catalogue no. ABIN101961)) and incubated at room temperature. pA-Tn5 was bound to antibodies, and transposition was performed at 37â°C and stopped using TAPS-Wash solution. Nuclei were lysed and pA-Tn5 decrosslinked using SDS-release solution. PCR was performed using KAPA HiFi plus dNTPs (Roche Diagnostics, catalogue no. 07958846001) with the following PCR settings: 72â°C 5âmin, 98â°C 30âs, 15 cycles of 98â°C 10âs, 63â°C 30âs, and 72â°C final extension for 1âmin, hold at 4â°C.
ATACâseq
Beads with nuclei were resuspended in ATACâseq solution (10âmM TAPS pH 8.5, 5âmM MgCl2, 10% DMF (Sigma-Aldrich, catalogue no. D4551), 0.2âµgâµlâ1 transposase (Tn5)) and incubated at 37â°C for 30âmin. Thereafter, 100âµl of DNA binding buffer (Zymo Research, catalogue no. D4003-1) was added and samples were stored at â20â°C. Then, DNA was extracted using Zymo DNA Clean and Concentrator-5 (Zymo Research, catalogue no. D4004). Library amplification was performed using KAPA HiFi plus dNTPs (Roche Diagnostics, catalogue no. 07958846001) and the following PCR settings: 72â°C 5âmin, 98â°C 30âs, 10 cycles of 98â°C 10âs, 63â°C 30âs, 72â°C 1âmin, hold at 4â°C.
Both ATACâseq and CUT&Tag libraries were cleaned using SPRI beads, eluted in nuclease-free water and pooled equimolecularly after library quantification using the KAPA library quantification kit (Roche Diagnostics, catalogue no. 079602). Libraries were sequenced in PE150 mode on a NovaSeq 6000 at Novogene.
Sequencing data processing
snRNA-seq data processing and analysis
Data integration and differential expression analysis for mouse snRNA-seq
The 10x Genomics Cell Ranger v.6.1.2 pipeline was used for demultiplexing, read alignment to reference genome mm10-2020A (10x Genomics), barcode processing and unique molecular identifier (UMI) counting with Include introns argument set to âTrueâ. The R package Seurat v.4.1.0 (ref.â76) was used to process, integrate and analyse datasets. scDblFinder77 was used to identify and remove doublets. Nuclei with unique feature counts less than 500 or greater than 3,000 and UMI counts greater than 40,000 were discarded during quality control (Extended Data Fig. 11a). Highly expressed genes such as mitochondrial genes, pseudogenes and Malat1 were excluded from the count matrix before normalization. SoupX78 was used to estimate potential ambient RNA contamination in all samples, but no sample required any correction. Samples were normalized using sctransform and integrated using the CCA (canonical correlation analysis) method built into Seurat. Filtered, normalized and integrated nuclei data were clustered by using the Louvain algorithm with a resolution of 0.4 using the first 30 principal components. Cluster markers were identified on the basis of differential gene expression analysis (Wilcoxon rank-sum test with |log2FC|â>â0.25 and adjusted Pâ<â0.05). Clusters were then annotated on the basis of known markers from literature34,36,37,46,79,80. Additionally, our manual cluster annotation was confirmed by reference mapping against a reference male mouse epiAT34 dataset (Extended Data Fig. 11b,c). Differential expression analysis (Wilcoxon rank-sum test with |log2FC|â>â0.5 and adjusted Pâ<â0.01) per cell type between different conditions was done using the FindMarkers function from Seurat. Differential expression analysis hits were intersected with a list of epigenetic modifier genes (see the Source Data to Extended Data Fig. 8) to investigate their expression dynamics. For visualization of snRNA-seq data we used the R package SCpubr v.1 (ref.â81).
Data integration and differential expression analysis for human snRNA-seq
The 10x Genomics Cell Ranger v.7.2.0 pipeline was used for demultiplexing, read alignment to reference genome GRCh38-2020-A (10x Genomics), barcode processing and UMI counting, with force cells set to 10,000. The R package Seurat v.4.1.0 (ref.â76) was used to process, integrate and analyse datasets. scDblFinder77 was used to identify and remove doublets. Nuclei with unique feature counts <300 or >4,000 (LTSS) / 6,000 (NEFA), UMI counts >15,000 (LTSS) / 25,000 (NEFA) and mitochondrial gene counts greater than 5% were discarded during quality control (Extended Data Fig. 12). SoupX78 was used to estimate and correct for potential ambient RNA contamination in all samples. Samples were normalized using sctransform and integrated using the CCA method built into Seurat. Filtered, normalized and integrated nuclei data were clustered by using Louvain algorithm using the first 30 principal components. For each study, the cluster resolution was determined using the R package clustree82. Cluster markers were identified on the basis of differential gene expression analysis (Wilcoxon rank-sum test with |log2FC| > 0.25 and adjusted Pâ<â0.01). Clusters were then annotated on the basis of known markers from literature34,35,36,37,83. Additionally, our manual cluster annotation was confirmed by reference mapping against reference human white AT atlas34 (Extended Data Figs. 2 and 3). For each AT depot, adipocytes from two studies were integrated together using the first 20 principal components following the steps as mentioned above. Differential expression analysis (Wilcoxon rank-sum test with |log2FC| > 0.5 and adjusted Pâ<â0.01) per cell type between different conditions was done using the FindMarkers function from Seurat. Differential expression analysis hits were validated using MAST and likelihood-ratio tests using the FindMarkers function from Seurat. For visualization of snRNA-seq data, we used the R package SCpubr v.1 (ref.â81).
SNP-based demultiplexing of human snRNA-seq datasets
To perform SNP calling and demultiplexing on the pooled samples, cellsnp-lite84 was first used to call SNPs on a cell level using the 1000 Genomes-based reference variant call file for hg38 at a resolution of 7.4âmillion SNPs. SNPs with less than 20 counts and a minor allele frequency of less than 10% were filtered out, as per the developer recommendations. Finally, the tool vireo85 was used to demultiplex the pooled data using the cellsnp-lite-derived genotype information.
For each donor, we analysed tissue composition and removed nuclei belonging to donors in the case in which no nuclei were assigned as adipocytes (one case in NEFA) or more than 50% or nuclei were assigned as B cells (one case in MTSS; lean donor) after correspondence with surgeons.
Transcriptional retention
DEGs from obese and WL cells from mouse and human were overlayed, respectively. A DEG was considered restored if it was no longer deregulated in WL cells when compared with controls. If not restored, we considered a DEG part of a transcriptional memory. Clusters identified as similar cell types (for example, three clusters of endothelial cells) were merged for DEG quantification but not differential expression analysis itself. For human snRNA-seq, only cell types for which we obtained at least 30 cells per donor were considered for the retention analysis. T cells were not included in differential expression analysis or transcriptional retention analysis. For integrated human adipocyte differential expression analysis quantification, non-coding transcripts were excluded.
TRAPâseq
Quality control of the raw reads was performed using FastQC v.0.11.9. Raw reads were trimmed using TrimGalore v.0.6.6 (https://github.com/FelixKrueger/TrimGalore). Filtered reads were aligned against the reference mouse genome assembly mm10 using HISAT2 v.2.2.1. Raw gene counts were quantified using the featureCounts86 program of subread v.2.0.1. Differential expression analysis was performed using the R package EdgeR87, with |log2FC| ⥠1 and nominal Pâ<â0.01 as cut-offs.
CUT&Tag and ATACâseq data processing and analysis
Quality control of CUT&Tag and ATACâseq data and generation of bedgraph files was performed as described previously75. Peaks were called from CUT&Tag sequencing and ATACâseq libraries on individual bedgraph files using SEACR88 v.1.3 in stringent mode with a peak calling threshold of 0.01. Peaks overlapping with mouse blacklist regions89 were filtered out. Called peaks were annotated using the R package ChIPSeeker90. Peak fold enrichment against genomic features was calculated using the formula: Σ(base pair (bp) overlap)âÃâgenome_size/[Σ(bp hPTM peak)âÃâΣ(bp genomic feature)]. Genomic features tracks were downloaded from ENCODE using the R package annotatr91. Visual quality control of bam files was performed with Seqmonk92. Called peaks were combined to generate a union peak list and quantified using the R package chromVAR93 v.1.16, generating a raw peak count matrix.
MOFA
MOFA50,94 was run to identify the driving variation source across all conditions using all data modalities. For each modality, the top 3,000 variable features (genes or peaks) between all samples were selected using the R package DESeq2 (ref.â95) and used as input to train the MOFA model. The trained MOFA model represented data variability in terms of five latent factors, which were further explored and visualized.
Generation of enhancer tracks of adipocytes
Adipocyte chromatin states were identified using ChromHMM v.1.22 (ref.â96) in concatenated mode with binned bam files (200-bp bins) from each condition combining all hPTMs and ATACâseq. After final model selection75 with eight chromatin states and emission parameter calculation of hPTMs and ATACâseq, chromatin state fold enrichment was performed against genomic features and ENCODE candidate cis-regulatory elements. Enhancer states were selected on the basis of genomic localization and hPTM enrichment. Subsequently, an enhancer track was generated per condition and merged for differential analysis.
Differential analysis of hPTMs and ATACâseq
Promoters
Promoters were defined using the getPromoters function from ChIPSeeker with TxDb.Mmusculus.UCSC.mm10.knownGene as input and setting the TSSRegion to c(-2000, 2000). Peaks overlapping with promoters were extracted using the annotatePeak function from ChIPseeker90 by selecting peaks annotated as promoters. For differential analysis, our raw peak count matrix was filtered for these promoter regions and counts were aggregated at gene level. Differential analysis of the same hPTM between two conditions was performed using the R package EdgeR87 with nominal Pâ<â0.01 and |log2FC|â>â1 as cut-offs.
Enhancers
ChromHMM was used to identify regions in the genome that were marked by H3K4me1, H3K27ac and open (ATACâseq) but not enriched for H3K4me3 and that were not promoters (Extended Data Fig. 9bâe). States 6 and 5 were selected as enhancer regions on the basis of their genomic locations (distal enhancer elements) (Extended Data Fig. 9bâe).
Our raw peak count matrix was filtered for enhancer regions defined by chromHMM, and peaks around the TSS (±2,000âbp) were discarded. Linkage of putative enhancers to genes was done using the R package ChIPSeeker by selecting the closest gene (TSS or gene body) within 20,000âbp distance. Putative enhancers farther away than 20,000 from a TSS or gene body were not linked to any gene and were discarded from downstream GSEA.
For each hPTM, the raw filtered peak matrices were log-normalized using the R package EdgeR and Pearsonâs correlation coefficient was computed using the cor function from the R package stats v.3.6.2.
Differential analysis of the same hPTM between two conditions was performed using the R package EdgeR with nominal FDRâ<â0.05 and |log2FC|â>â1 as cut-offs.
PCA
Raw gene and promoter/enhancer-specific peak count matrices were log-normalized using the R package EdgeR. PCA of the normalized count matrices was performed using the prcomp function of R package stats v.3.6.2.
GSEA
GSEA was performed using the R package enrichR97,98,99. For generation of heatmaps summarizing GSEA across cell types, significantly enriched terms were selected using the adjusted P value (<0.01) and the combined.score (enrichment score) was scaled and visualized.
Visualization
R v.4.2, GraphPad Prism v.9.5.1 and Seqmonk v.1.48.1 were used to generate plots and Affinity Designer and Publisher were used to adjust plots for clarity (for example, colour schemes).
Statistical analysis of physiological parameters from mice
GraphPad Prism v.9.5.1 was used to analyse physiological data from mice. Each dataset of physiological parameters was tested for normality using the ShapiroâWilk test. On the basis of the results, parametric or non-parametric tests were used to compare experimental with age-matched control groups. Tests are indicated in figure legends and the Source Data.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.