Ethics
Fresh tumour samples were obtained by surgery from patients at Paul Brousse and Institut Curie care centres. All patients provided written informed consent for use of tumour samples. The study was approved by institutional regulatory boards (no. 587 and DATA190160). All cLip-1 in vivo experiments were performed in compliance with the German Animal Welfare Law and were approved by the Institutional Committee on Animal Experimentation and the Government of Upper Bavaria (no. ROB-55.2-2532.Vet_02-18-13). All intranodal injection mouse experiments complied with all relevant ethical regulations and were performed according to protocols approved by the Institutional Animal Care and Use Committee at Harvard T. H. Chan School of Public Health (protocol IS00003460). For mouse lymph and blood collection, animal experiments were performed in accordance with the European Community guidelines for the care and use of animals. Animal experiments were performed in agreement with the French guidelines for animal handling and approved by local ethics committees (agreement no. 16487-2018082108541206 v3).
Chemical synthesis
Starting materials were purchased at the highest commercial quality and used without further purification unless otherwise stated. Anhydrous solvents were obtained by passing the degassed solvents through molecular sieves and activated alumina columns. Reactions were monitored by thin layer chromatography (TLC) using aluminium plates coated with silica gel or neutral aluminium oxide from Merck (60 F254). TLC plates were visualized by UV or by treatment with a ninhydrin, ceric ammonium molybdate or potassium permanganate solution and heating. Reaction products were purified by flash column chromatography on silica gel 60 (230–400 mesh, Macherey Nagel) or aluminium oxide (activated neutral, Sigma-Aldrich) using a CombiFlash NextGen System and a preparative HPLC Quaternary Gradient 2545 equipped with a photodiode array detector 2998 (Waters) fitted with a reverse-phase column (XBridge BEH C18 OBD prep column 5 μm, 30 × 150 mm). NMR spectroscopy was performed using Bruker 400 or 500 MHz instruments. Spectra were run in methanol-d4, dimethylsulfoxide-d6, methylene chloride-d2 or chloroform-d at 298 K or 310 K as indicated. 1H chemical shifts δ are expressed in ppm using the residual non-deuterated solvent as an internal standard, and the coupling constants J are specified in Hz. The following abbreviations are used: bs, broad singlet; s, singlet; d, doublet; dd, doublet of doublets; ddd, doublet of doublet of doublets; dt, doublet of triplets; dq, doublet of quadruplets; q, quadruplet; t, triplet; td, triplet of doublets, quint., quintet; and m, multiplet. 13C chemical shifts δ are expressed in ppm using the residual non-deuterated solvent as an internal standard. The purity of the final compounds was determined to be >98% by UPLC–MS. Low-resolution mass spectra were recorded using a Waters Acquity H-class equipped with a photodiode array detector and a SQ Detector 2 (UPLC-MS) fitted with a reverse-phase column (Acquity UPLC BEH C18 1.7 μm, 2.1 × 50 mm). High-resolution MS spectra were recorded on a Thermo Scientific Q-Exactive Plus equipped with a Robotic TriVersa NanoMate Advion. Procedures for the synthesis of small molecules are detailed in the Supplementary Information.
NMR titration experiments
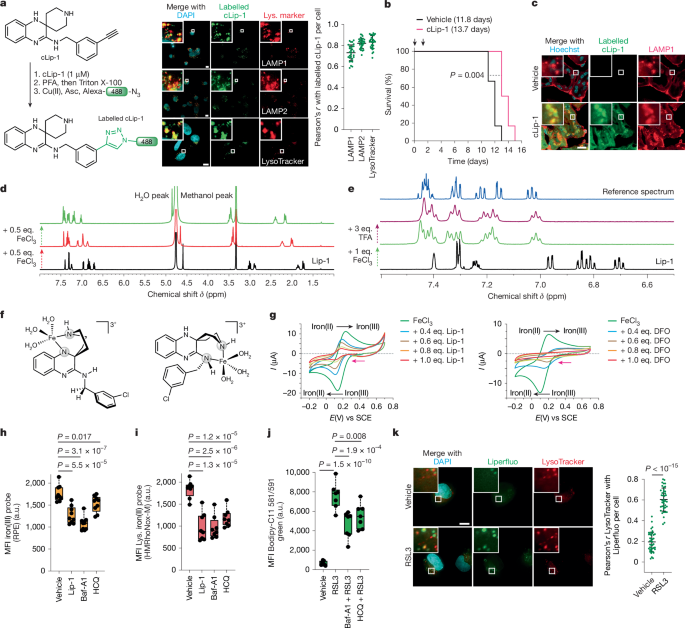
1H NMR spectra were recorded on a Bruker 500 MHz spectrometer at 310 K using Bruker Topspin (v.4.1.4) software and analysed using MestRenova (v.5.0.1-35756) or Bruker Topspin (v.4.1.4) software, and chemical shifts δ are expressed in ppm using the residual non-deuterated solvent signals as an internal standard. All the solutions were prepared in methanol-d4. For recordings of 1H NMR spectra of Lip-1 titrated with FeCl3 before the addition of TFA or sodium deuteroxide, 0.1–1 eq. of a solution of FeCl3 (Alfa Aesar, 12357, 2 µl, 145 mM) was added to a solution of Lip-1 (Sigma-Aldrich, SML1414, 1 mg in 600 µl). Then, a solution of TFA (Sigma-Aldrich, T6508, lot STBG1988V, 20 µl, 3 eq. 438 mM) or a solution of sodium deuteroxide (Eurisotop, D076Y, lot R2621, 1 µl) was added. For recordings of 1H NMR spectra of Lip-1 and naphthalene titrated with FeCl3, 0–1 eq. of a solution of FeCl3 (3 µl, 97 mM) was added over a solution of Lip-1 (1 mg in 500 µl) and naphthalene (Sigma-Aldrich, 147141, 1 eq., 100 µl, 28.9 mM). After each addition, the solution was stirred for few seconds and NMR spectra were recorded.
Cyclic voltammetry
Cyclic voltammetry26 experiments were performed with a three-electrode cell. A saturated calomel electrode was used as reference, a steady glassy carbon electrode of diameter 3 mm was selected as the working electrode and a platinum wire as the counter-electrode. All cyclic voltammograms were recorded at room temperature with a μ-autolab III from Metrohm using Nova software (v.2.1) with a scan rate of 0.1 V s–1. HPLC-grade acetonitrile and methanol were used for recordings. For all experiments, nBu4NBF4 in acetonitrile (0.1 M, 32.9 mg ml–1 stock solution) was used. A 10 ml of a stock solution of 1 mM FeCl3 was prepared with 500 µl of 20 mM FeCl3 solution in milliQ water and 9.5 ml acetonitrile. Then, 5 µl (0.2 eq.) of a 40 mM stock solution of DFO, Lip-1, metcLip-1 (in-house) or alcLip-1 (in-house, solubilized in acetonitrile or methanol) was added to 1 ml of 1 mM FeCl3 solution until 1.0 eq. was reached. After each addition, the solution was stirred for few seconds and voltammograms were recorded.
Molecular modelling
All structures were optimized using the Gaussian 16 (ref. 50) set of programs at the BHLYP/SVP level for all atoms (sextet spin state for iron(III) complexes) (Gaussian 16, revision C.01). Thermal correction to the Gibbs free energy was computed at 310.15 K. Single points at the UMP2/SVP level were performed using the SMD solvation model (methanol). The results presented are ΔG310 in kcal mol–1.
Fluorimetry of the iron(II) probe RPE
A solution of RPE (in-house, 10 mM) in DMSO was diluted with a solution of acetonitrile–acetate buffer (10 mM, pH 5, 2:1, v/v) to reach a concentration of 10 µM. In a 96-well plate, three solutions of RPE (100 µl, 10 mM) and 10, 20 or 50 eq. of Lip-1 (in-house 1, 2 or 5 µl of 10 mM stock solution in methanol, respectively) were prepared and the solutions were mixed several times. Then, 10 eq. FeCl3 (1.0 µl of stock solution in acetonitrile–acetate buffer (10 mM, pH 5, 2:1, v/v)) was added to each solution and mixed again. Spectra were recorded with a fluorimeter (Tecan spark 10 M) (λex = 510 nm, λem = 540–700 nm) using SoftMax Pro 7.1 GxP software. UPLC-grade acetonitrile and methanol as well as milliQ water were used.
Fluorescence-enabled inhibited autoxidation (FENIX) assay
A conical centrifuge tube (15 ml) was charged with unilamellar liposomes of egg phosphatidylcholine (0.4 ml of a 20 mM suspension in PBS, 100 nm average diameter), 134 ml of a 12 mM solution of (E)-1,2-bis((2-methyldecan-2-yl)oxy)diazene (Cayman, 32742) in ethanol and PBS (7.46 ml of 12 mM at pH 7.4) and vortexed. STY-Bodipy (3.8 μl of a 1.74 mM solution in DMSO) was then added and the suspension vortexed once. The liposome–initiator–dye mixture was transferred to a reservoir, and using a 300 μl multichannel pipette, a 96-well plate (black, Nunc) was charged with the mixture (295 μl per well). Test compounds (5 μl of a 0.24 mM solution in acetonitrile) were added to bring the final volume to 300 μl. The contents of the wells were manually mixed using a multichannel pipette and then mixed in a microplate reader for 1 min at 37 °C, after which the oxidation of STY-Bodipy was monitored by fluorescence (λex/λem = 488/518 nm) overnight. Data were collected using Agilent BioTek Gen5 software (v.3.08.01).
Antibodies
Antibodies are annotated as follows: WB, western blot; FCy, flow cytometry; FI, fluorescence imaging; Hu, used for human samples; Ms, used for mouse samples. Dilutions are indicated. Any antibody validation by the manufacturer is indicated and can be found on the manufacturers’ websites. Our antibody validation knockdown (KD) and/or knockout (KO) strategies for relevant antibodies are indicated. The following primary antibodies were used: AIFM2/FSP1 (Merck, MABC1638, clone 6D8-11, lot Q3745998, WB, 1:500, Hu, KD validated in-house); catalase (Cell Signaling, 12980T, clone D4P7B, lot 3, FI, 1:200, Hu); CD3-BV510 (BioLegend, 317332, clone OKT3, lot B263750, FCy, 1:100, Hu); CD31-PE-Cy7 (BioLegend, 303118, clone WM59, lot B276836, FCy, 1:100, Hu); CD31-BV605 (BioLegend, 303122, clone WM59, lot 331683, FCy, 1:100, Hu); CD31-BV605 (BioLegend, 102427, clone 390, lot B375532, FCy, 1:100, Ms); CD44 (Abcam, ab189524, clone EPR18668, lot 1014086-32, WB, 1:30,000, Hu, KO and KD validated in-house); CD44-AF647 (Novus Biologicals, NB500-481AF488, clone MEM-263, lot P158343, FCy, 1:100, Hu); CD44-AF647 (Novus Biologicals, NB500-481AF647, clone MEM-263, lot D145771, FCy, 1:100, Hu); CD44-AF647 (BioLegend, 103018, clone IM7, lot B317762, FCy, 1:100, Ms); CD45-BV785 (BioLegend, 304048, clone HI30, lot B339809, FCy, 1:100, Hu); CD45-BV510 (BioLegend, 368526, clone 2D1, lot B373428, FCy, 1:100, Hu); CD45-BV510 (BioLegend, 103138, clone 30-F11, lot B386738, FCy, 1:100, Ms); CD163-PerCP/Cyanine5.5 (BioLegend, 326512, clone RM3/1, lot B291202, FCy, 1:100, Hu); COXIV (Abcam, ab16056, lot GR3206555-1, WB, 1:1,000, Hu); cytochrome c (Cell Signaling, 12963S, clone 6H2.B4, lot 2, FI, 1:200, Hu); EEA1 (Abcam, ab70521, clone 1G11, lot GR315680-1, FI, 1:200, Hu, validated by immunocytochemistry/immunofluorescence by manufacturer); FAP-AF700 (R&D Systems, FAB3715N, clone 427819, lot AEVI020011, FCy, 1:100, Hu); FAP-AF750 (Bio-Techne, FAB3715S-100UG, clone 427819, lot 1718688, FCy, 1:100, Hu); ferritin (Abcam, ab75973, clone EPR300AY, lot 10136442-29, WB, 1:1,000, Hu, validated by WB by manufacturer); fibronectin (Abcam, ab45688, clone F14, lot 1016266-35, WB, 1:1,000, Hu); FTH1 (Santa Cruz Biotechnology, sc-376594, clone B-12, lot G2622, WB, 1:200, Hu); GPX4 (Abcam, ab125066, clone EPNCIR144, lot GR3369574-4, WB, 1:2,000, Hu, KO validated by manufacturer, KD validated in-house); 4-HNE (Abcam, ab48506, clone HNEJ-2, lot 1062274-2, FI, 1:200, Hu); IRP2 (Novus Biologicals, NB100-1798, lot D-4, WB, 1:1,000, Hu); Lamin A/C (Cell Signaling, 2032S, lot 6, WB, 1:1,000, Hu); LAMP1 (Cell Signaling, 9091S, clone D2D11, lot 7, FI, 1:200, Hu); LAMP1 (Santa Cruz Biotechnology, sc-20011, clone H4A3, FI, 1:100, Ms); LAMP1 (Abcam, ab24170, lot GR3235630-1, WB, 1:1,000, Hu); LAMP2 (Abcam, ab25631, clone H4B4, lot 1011336-1, FI, 1:200, Hu); LAMP2 (Thermo Fisher Scientific, MA1-205, clone H4B4, lot YG377512, WB, 1:1,000, Hu); LIMPII (Proteintech, 27102-1-AP, WB, 1:1,000, Hu); NCOA4 (Abcam, ab86707, lot GR3244520-13, WB, 1:10,000, Hu, KD validated in-house); NDUFS1 (Abcam, ab157221, clone EPR11522(B), lot YJ110907DS, WB, 1:1,000, Hu); PDIA3 (Sigma-Aldrich, AMAB90988, clone CL2444, lot 02879, FI, 1:200, Hu; WB, 1:1,000, Hu); MHCII-APC/Cyanine7 (BioLegend, 107628, clone M5/114.12.2, lot B370049, FCy, 1:100, Ms); RCAS1 (Cell Signaling, 12290S, clone D2B6N, lot 1, FI, 1:200, Hu, WB, 1:1,000, Hu); SLC7A11/xCT (Cell Signaling, 12691S, clone D2M7A, lot 5, WB, 1:1,000, Hu, KD validated in-house); SOX4 (Santa Cruz Biotechnology, sc-518016, clone B-7, lot G2023, WB, 1:200, Hu); TFR1 (Life Technologies, 13-6800, clone H68.4, lot VJ313549, WB, 1:1,000, Hu, KO and KD validated in-house); TFR1-APC-AF750 (Beckman Coulter, A89313, clone YDJ1.2.2, lot 200060, FCy, 1:100, Hu); TFR1-PE (BioLegend, 334106, clone CY1G4, lot B364886, FCy, 1:100, Hu); γ-tubulin (Sigma-Aldrich, T5326, clone GTU-88, lot 0000140390, WB, 1:1,000, Hu, validated by manufacturer); and vimentin (Cell Signaling, 5741S, clone D21H3, lot 8, WB, 1:1,000, Hu). The following secondary antibodies were used: Alexa Fluor 647 anti-mouse (Abcam, ab150115, tissue labelling, 1:500, Ms); Alexa Fluor 647 anti-mouse (Invitrogen, A21237, lot 1485202, FI, 1:1,000, Hu); Alexa Fluor 647 anti-rabbit (Invitrogen, A21246, lot 2714437, FI, 1:1,000, Hu); donkey anti-rabbit IgG-H+L HRP-conjugated (Bethyl Laboratories, A120-108P, lot 13, WB, 1:10,000, Hu); goat anti-mouse IgG-H+L HRP-conjugated (Bethyl Laboratories, A90-116P, lot 39, WB, 1:10,000, Hu); and goat anti-rat IgG-H+L HRP-conjugated (Invitrogen, 31470, WB, 1:10,000, Hu).
Cell culture
HT-1080 cells, MDA-MB-231 and 4T1 cells were obtained from the ATCC. Dissociated human and mouse tumour cells and 4T1 cells were cultured in RPMI 1640 supplemented with GlutaMAX (Gibco, 61870010) and 10% FBS (Eurobio Scientific, CVFSVF00-01). HT-1080 cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM)–GlutaMAX (Gibco, 61965059) supplemented with 10% FBS (Gibco, 10270-106) and penicillin–streptomycin (BioWhittaker/Lonza, DE17-602E). MDA-MB-231 cells were cultured in DMEM–GlutaMAX supplemented with pyruvate (Thermo Fisher Scientific, 31966021) and supplemented with 10% FBS and penicillin–streptomycin (Gibco, 15140148). FC1242 and FC1245 mouse pancreatic cancer cells, 4a cells and human pancreatic hMIA-2D cells were a gift from the Tuveson laboratory (Cold Spring Harbor Laboratory) and were cultured in DMEM–GlutaMAX supplemented with 10% FBS and penicillin–streptomycin. Primary human pancreatic PDAC024T, PDAC030T, PDAC053T, PDAC054T, PDAC084T, PDAC090T and PDAC211T cells were grown in serum-free ductal medium DMEM/F12 (Gibco, 10565018) supplemented with 0.61 g in 500 ml nicotinamide (Sigma-Aldrich, 3376), 2.50 g in 500 ml glucose (Sigma-Aldrich, G6152), 1:200 ITS+ (Corning, 354352), 1:20 Nu-serum IV (Corning, 355104), 100 ng ml–1 cholera toxin, 1 µM dexamethasone (Sigma-Aldrich, D4902), 50 nM 3,3′,5-triiodo-l-thyronine (Sigma-Aldrich, T6397) and penicillin–streptomycin). Fresh bovine pituitary extract (Gibco, 13028-014, 22.7 ng ml–1) and 50 ng ml–1 animal-free recombinant human EGF (Thermo Fisher Scientific, AF-100-15-1MG) were added to new medium when cells were split or plated. SUM159 cells were cultured in Ham F12, GlutaMAX (Gibco, 31765035), NEAA (Gibco, 11140050), antibiotic–antimycotic (Thermo Fisher Scientific, 15240062), insulin (Humalog, Cip: 3400934142680), hydrocortisone (Sigma-Aldrich, H0888-10G) and 5% FBS (Thermo Fisher Scientific, A5256701). Primary lung circulating tumour cells (Celprogen, 36107-34CTC, lot 219411, sex: female) and primary colon circulating tumour cells (Celprogen, 36112-39CTC, lot 20188, sex: female) were grown using stem cell complete medium (Celprogen, M36102-29PS) until the third passage. Circulating cancer cells were grown in stem cell ECM T75-flasks (Celprogen, E77002-07-T75). Pfa1 cells were cultured using DMEM high glucose and 10% FBS, 2 mM l-glutamine and 1% penicillin–streptomycin.
Dissociation of human and mouse tumour samples
Tumour samples were collected from patients after surgery. Tumour samples were from patients with PDAC, UPS, liposarcoma, angiosarcoma, epithelioid sarcoma or PDAC liver metastasis. Tumours were dissociated using a human tumour dissociation kit (Miltenyi, 130-095-929) according to the manufacturer’s protocol. In brief, tumours were cut into small pieces (1–5 mm), put into the enzyme mix in RPMI and dissociated using gentleMACS Octo Dissociator with Heaters (Miltenyi) with the appropriate gentleMACS program (37C_h_TDK). Per tumour sample, 9.4 ml RPMI medium was used with the corresponding enzyme concentration according to the manufacturer’s protocol. Mouse tumour samples were dissociated using a mouse tumour dissociation kit (Miltenyi, 130-096-730) according to the manufacturer’s protocol. Tumours were cut into small pieces (1–5 mm), put into the enzyme mix in RPMI and dissociated using gentleMACS Octo Dissociator with Heaters (Miltenyi) with the appropriate gentleMACS program (37C_m_TDK). Subsequently, the dissociated tumour suspension was applied to a MACS SmartStrainer (30 µm) (Miltenyi). Samples were diluted with 1× PBS and centrifuged at 300g. The cell pellet was resuspended in RPMI (with 10% FBS and penicillin–streptomycin), and cells were counted using an automated cell counter (Entek). The total population of dissociated cells is denoted as dissociated tumour cells, whereas the subpopulation corresponding to cancer cells from tumours by negative selection (see the section ‘Flow cytometry’) is denoted as dissociated tumour cancer cells.
Establishment of xenograft-derived primary cell cultures
These models were originally derived from patient-derived xenograft (PDX) models. The PDX fragments designated for cell culture were processed in a biosafety chamber. After mincing to small pieces, they were treated with collagenase type V (Sigma-Aldrich, C9263) and trypsin–EDTA (Gibco, 25200-056) and suspended in DMEM supplemented with 1% w/w penicillin–streptomycin and 10% FBS. After centrifugation, cells were resuspended in serum-free ductal medium adapted from previous protocols51 at 37 °C in a 5% CO2 incubator. Amplified cells were stored in liquid nitrogen. Cells were weaned from antibiotics for more than 48 h before testing. This protocol was used to establish the cells designated as PDAC024T, PDAC030T, PDAC053T, PDAC054T, PDAC084T, PDAC090T and PDAC211T.
Establishment of xenograft-derived pancreatic organoids
Xenograft-derived pancreatic organoid (XDPO) models were originally derived from PDX models. Xenografts were split into several small pieces and processed in a biosafety chamber. After mincing to small pieces, they were treated using a human tumour dissociation kit (Miltenyi, 130-095-929). Undigested pellets were digested with accutase (Thermo Fisher Scientific, A1110501) at 37 °C for 30 min. The pancreatic tissue slurry was transferred to a 100 μm tissue strainer and then placed into a 12-well plate coated with 150 μl GFR Matrigel (Corning, 354230). The samples were cultured in pancreatic organoid feeding medium, which consisted of advanced DMEM/F12 supplemented with 10 mM HEPES (Thermo Fisher Scientific, 15630056), 1× GlutaMAX (Thermo Fisher Scientific, 35050087), penicillin–streptomycin, 100 ng ml–1 animal-free recombinant human FGF10 (Thermo Fisher Scientific, 500-P151G-50UG), 50 ng ml–1 animal-free recombinant human EGF (Thermo Fisher Scientific, AF-100-15-1MG), 100 ng ml–1 recombinant human noggin (Bio-Techne, 6057-NG), WNT3A-conditioned medium (30% v/v), RSPO1-conditioned medium (10% v/v), 10 nM human gastrin 1 (Sigma-Aldrich, SCP0152), 10 mM nicotinamide (Sigma-Aldrich, N0636), 1.25 mM N-acetylcysteine (Sigma-Aldrich, A9165), 1× B27 (Thermo Fisher Scientific, 17504001), 500 nM A83-01 (Tocris, 2939/10) and 10.5 μM Y27632 (Tocris, 1254/1). The plates were incubated at 37 °C in a 5% CO2 incubator, and the medium was changed every 3–4 days. This procedure was used to generate the XDPOs PDAC009T, PDAC003T, PDAC117T and PDAC372T.
Chemosensitivity profiling of XDPOs
For chemosensitivity profiling, XDPOs were plated into 96-well plates and then subjected to incrementally increasing concentrations of drugs. Cell viability was measured 72 h after treatment using CellTiter-Glo 3D (Promega, G9683). Doubling times of XDPO viability for vehicle-treated conditions were calculated on days 0 and 3. The ratio of day 3 over day 0 corresponded to the replication rate (RR) of the cells at 72 h. Doubling times were calculated using the formula 72 × 2/RR. Fluorescence and luminescence values were quantified using a Tristar LB941 plate reader (Berthold Technologies). Each experiment was performed at least three times with at least three replicates.
Cell death and viability assays
Cell death assay with Annexin-V and propidium iodide
PDAC053T cells were seeded in 6-well plates at a density of 2 × 105 cells per well. RSL3 (Sigma-Aldrich, SML2234, 0.1, 0.5, 2 and 10 μM) was added together with Lip-1 (1 µM), cLip-1 (1 µM), alcLip-1 (10 µM) or metcLip-1 (10 µM) on the following day. After 24 h, the medium was recovered and cells were trypsinized. Cells were collected, pelleted along with the recovered medium and washed with 1× PBS. Next, 100 μl of 1× Annexin-V binding buffer containing Annexin-V and propidium iodide was added according to the manufacturer’s protocol (Annexin-V flow cytometry kit, Thermo Fisher Scientific, V13242). 1× PBS buffer containing 10% FBS and EDTA (0.1% v/v) was added for flow cytometry. Flow cytometry was performed using an AttuneTM NxT flow cytometer and data were analysed using FlowJo.
Cell death assay with Annexin-V and Sytox blue
PDAC053T cells were seeded in 6-well plates at a density of 2 × 105 cells per well. The following day, cells were transfected with siRNA as detailed in the section ‘RNA interference’. Medium was replaced 6 h after transfection. Three days after transfection, cells were treated with the indicated concentrations of Fento-1 for 6 h. Cell death was analysed using Annexin-V AF488 (Thermo Fisher Scientific, A13201) and Sytox blue (Thermo Fisher Scientific, S34857). Flow cytometry was performed using an AttuneTM NxT flow cytometer and data were analysed using FlowJo.
LDH assay
LDH release was measured using a Cytotoxicity detection kit (Sigma-Aldrich, 11644793001) according to the manufacturer’s protocol in a 96-well plate. Cell viability was assessed using a CellTiterGlo 2.0 (Promega, G9241) or CellTiter blue (Promega, G8081) kit according to the manufacturer’s protocol in a 96-well plate. In brief, 4,000 cells (HT-1080, PDAC053T or 4T1) were seeded per well in clear-bottom and darkened 96-well plates (Greiner, 655090, lot E23063EG) 24 h before the experiment. Cells were then pretreated for 2 h with Lip-1 (10 µM), cLip-1 (10 µM), ferrostatin-1 (SML0583, 10 µM), DFO (Sigma-Aldrich, D9533, 100 µM), deferasirox (Cayman chemical, 16753, 10 µM), deferiprone (Sigma-Aldrich, Y0001976, 100 µM), α-tocopherol (100 µM), vitamin K3 (Sigma-Aldrich, M5625-25G, 10 µM), Z-VAD-FMK (Enzo Life Sciences, ALX-260-020-M005, 50 µM) or necrostatin-1 (Sigma-Aldrich, N9037, 20 µM). Subsequently, Fento-1 (10 µM, 6 h) was added. For dose–response measurements, cells were treated with varying amounts of the respective molecule for the time indicated. Samples were processed as detailed in the manufacturer’s protocols and data were recorded on a SpectraMax ID3 plate reader (Molecular Devices). For standard-of-care cell-viability measurements, cells were plated at 2,000 cells per well 24 h before the experiment. Cells were incubated with serial dilutions of Fento-1, irinotecan (Sigma-Aldrich, I1406), 5-fluorouracil (5-FU; Alfa Aesar, A13456-06) or oxaliplatin (Bio-Techne, 2623) for 72 h.
MTT assay
Cells were plated in a 96-well plate, incubated for 24 h and then pretreated with Lip-1 (1 µM) for 10 min before treatment with vehicle control or Fento-1 for 24 h. After 24 h, the medium was carefully removed and 50 μl serum-free medium and 50 μl MTT solution (Cayman Chemical, 21795) were added per well. The plate was incubated at 37 °C for 3 h. After incubation, the solution was removed and 100 μl DMSO was added per well. The plate was covered and shaken on an orbital shaker for 15 min before reading the absorbance on a plate reader (OD = 590). For RSL3 treatment, Pfa1 cells were seeded in 96-well plates (2,000 cells per well) and cultured overnight. The following day, cells were treated with RSL3 (500 nM) and the indicated small molecules in serial dilution. For 4-OH-TAM treatment, Pfa1 cells were seeded in 96-well plates (500 cells per well) with 4-OH-TAM (1 μM) and a dilution series of the indicated compounds. After 24 h (for RSL3) or 72 h (for 4-OH TAM) incubation, cell viability was assessed using resazurin as a cell-viability indicator. Fluorescence intensity was measured at λex/λem = 540/590 nm using a SpectraMax iD5 microplate reader with SoftMax Pro v.7 software (Molecular Devices) after 4 h of incubation in standard cell culture medium containing 0.004% resazurin.
Alamar blue assay
The effect of sublethal doses of Fento-1 together with RSL3 or ML210 on cell viability was assessed using the Alamar blue assay. To determine cell viability, 3,000 HT-1080 or MDA-MB-231 cells were seeded in a 96-well plate with standard medium (10% FBS and 1% penicillin–streptomycin) around 20 h before treatment. Cells were pretreated with 0.5 µM Fento-1 and/or 0.5 µM Lip-1 for 1 h. After 48 h of RSL3 or ML210 (Sigma-Aldrich, SML0521-5MG) treatment, a cell-viability assay was performed. Alamar blue solution was made by dissolving 0.5 g resazurin sodium salt (Sigma-Aldrich, 263-718-5) in 100 ml sterile PBS and sterile filtrated through a 0.22 µm filter. After 2 h of incubation, viability was assessed by measuring the fluorescence using a 540 ± 20 nm excitation filter and a 590 ± 20 nm emission filter on a Spark microplate reader (Tecan).
IncuCyte measurements of the cytotoxicity of Fento-1
Kinetics of cell death were collected using an IncuCyte bioimaging platform (Essen). Cells were seeded in 96-well plates (3,000 cells per well) 1 day before treatment. Cells were treated with Fento-1 in combination with Lip-1 (0.5 µM) in FluoroBrite DMEM medium (Thermo Fisher Scientific A1896701). Four images per well were captured every hour, analysed and data were averaged. Cell death was measured on the basis of the incorporation of DRAQ7 (0.1 µM, Cell Signaling, 7406S). Data were collected as count of DRAQ7-positive cells per total number of cells in each condition.
Generation of HT-1080 DTP cancer cells
About 2 × 106 HT-1080 cells were plated in T75 flasks 24 h before adding doxorubicin (Clinisciences, HY-15142, 25 nM). Flasks were washed with 1× PBS buffer, and medium containing doxorubicin (25 nM) was replaced every 3–4 days. After 30 days of treatment with doxorubicin, 4,000 cells were plated in 96-well plates per well to assess cell viability after treatment with Fento-1.
Clonogenic survival analysis
About 1 × 106 SUM159 cells were plated in 10 cm dishes in complete growth medium. Seeded cells were treated with doxorubicin at the half maximal inhibitory concentration (IC50) concentration (150 nM). After 72 h, cells were washed and treated with Fento-1 (5 µM, 72 h) or 0.2% DMSO without drug. After 72 h, cells were washed, and the treatment was replaced with complete growth medium. After 10–15 days, cells were washed, fixed and stained with acetic acid and methanol (1:7) and 1% Coomassie blue 1 h at room temperature. The number of colonies with >100 cells was counted using a SCAN1200 colony counter (Interscience).
Drug synergy analysis
Cells were plated in 96-well plates (5,000 cells per well). After 12 h, cells were treated with 5-FU (Selleckchem, S1209, 5 µM), gemcitabine (Selleckchem, S1714, 1 µM), oxaliplatin (Selleckchem, S1224, 20 µM), paclitaxel (Selleckchem, S1150, 0.1 µM) or SN-38 (Selleckchem, S4908, 0.1 µM) alone or in combination with Fento-1 (1.5 µM) for 72 h. Cell viability was estimated after the addition of PrestoBlue (Life Technologies, A13261) for 2 h following the manufacturer’s instructions. To determine the existence of a synergistic effect on cell proliferation or cell viability between standard-of-care drugs and Fento-1, we used the software SynergyFinder. For the analysis of this effect, the ZIP score was used as a model for calculating synergy52.
cLip-1 treatment in vivo
Mice were kept under standard conditions with water and food ad libitum and in a controlled environment (22 ± 2 °C, 55 ± 5% humidity, 12 h light–dark cycle). For animal studies, C57BL6/J mice were randomized into separate cages. Mice aged 12–24 weeks and sex-matched were used for all experiments. For the survival cohort study, Rosa26-creERT2;Gpx4f/f mice were intraperitoneally treated with tamoxifen (Sigma-Aldrich, T2859, 2 mg per day dissolved in Myglyol 812) on days 0 and 1 for deletion of Gpx4 in the whole body except in the brain2. From day 2, cLip-1 (in-house, 10 mg per kg per day dissolved in 1× PBS containing 20% PEG400 and 5% Solutol HS15) or vehicle was intraperitoneally administered to the mice each day until the completion of the survival study. For histochemistry analyses, Rosa26-creERT2;Gpx4f/f mice treated with intraperitoneal tamoxifen injection (2 mg per day on days 0 and 1) were used. On day 7, cLip-1 (in-house, 10 and 100 mg kg–1) or vehicle was intraperitoneally injected into the mice. At 1 h after the injection, mice were euthanized, and the kidney and liver samples were processed as described in the section ‘Small-molecule labelling using click chemistry’.
Isolation of blood, serum and lymphatic fluid from mice
Balb/c mice (25-week-old females) were purchased from Charles River and housed at the CRCM animal core facility and randomized into separate cages. Mice aged 12–24 weeks and sex-matched were used for all experiments. Mice were housed under sterile conditions with sterilized food and water provided ad libitum and maintained on a 12 h light–dark cycle and 22 ± 2 °C, 55 ± 5% humidity. Mice were not subjected to any procedures before collection of lymph, blood and serum.
For lymph sample collection, 30 min before the beginning of the experiment, buprenorphine (Buprecare), an analgesic, was administered by intraperitoneal injection (0.5 mg kg–1). Mice were euthanized by intraperitoneal injection of a ketamine–xylazine combination (ketamine 100 mg kg–1 (Imalgène) and xylazine 10 mg kg–1 (Rompum); 20 µl g–1). After cutaneous and peritoneal incisions, the lymph in the intestinal lymph trunk53 was collected using a glass capillary. Lymph samples were placed in cryotubes, frozen at −20 °C and stored at −80 °C.
For blood and serum sample collection, after lymph collection, we performed a terminal cardiac puncture (23 G needle with a 1–2 ml syringe) by thoracotomy to collect a large volume of blood without anticoagulants. Next, 100–200 µl of the blood sample was placed in a vial, frozen at −20 °C and stored at −80 °C. The rest of the blood sample was left to stand at room temperature for 30 min, then centrifuged at 2,000g for 15 min. The supernatant (serum) was collected in a vial, frozen at −20 °C and stored at −80 °C.
Quantitative proteomics
Sample preparation
HT-1080 cells were grown in 148 cm2 round dishes at 70% confluence and treated with Fento-1 (1 µM, 48 h). Whole-cell extracts were collected and then centrifuged at 500g for 5 min at 4 °C, washed 3× with ice-cold 1× PBS and lysed using lysis buffer (8 M urea, 200 mM sodium bicarbonate and complete protease inhibitor cocktail (Roche, 000000011697498001)). Lysis was achieved by incubating on a rotating wheel at 25 r.p.m. for 45 min at 4 °C. Lysates were centrifuged at 20,000g for 20 min at 4 °C, and the supernatant that contained total protein was used for quantitative global proteomic analysis. In brief, 30 µg total protein cell lysate was reduced by incubating with 5 mM dithiothreitol at 57 °C for 30 min and then alkylated with iodoacetamide (10 mM, 30 min) at room temperature in the dark. Trypsin/LysC (Promega) was added at 1:100 (w/w) enzyme to substrate. Digestion was performed overnight at 37 °C. Samples were then loaded onto custom-made C18 StageTips (AttractSPE Disk Bio C18-100.47.20, Affinisep) for desalting. Peptides were eluted using a ratio of 40:60 acetonitrile and H2O + 0.1% formic acid and vacuum concentrated to dryness with a SpeedVac. Peptides were reconstituted in 0.3% TFA before liquid chromatography–tandem mass spectrometry (LC–MS/MS).
LC–MS/MS analyses
LC was performed with a Vanquish Neo LC system (Thermo Scientific) coupled to an Orbitrap Astral mass spectrometer, interfaced by a Nanospray Flex ion source (Thermo Scientific). Peptides were injected onto a C18 column (75 µm inner diameter × 50 cm double nanoViper PepMap Neo, 2 μm, 100 Å, Thermo Scientific) regulated at 50 °C and separated with a linear gradient from 100% buffer A (100% H2O and 0.1% formic acid) to 28% buffer B (100% acetonitrile and 0.1% formic acid) at a flow rate of 300 nl min–1 over 104 min. The instrument was operated in data-independent acquisition (DIA) mode. MS full scans were recorded on the Orbitrap mass analyser in centroid mode for ranges 380–980 m/z with a resolution of 240,000, a normalized automatic gain control (AGC) target set at 500% and a maximum injection time of 5 ms. The DIAs in the Astral analyser were performed in centroid mode for a mass range of 380–980 m/z with a window width of 2 Da (without overlap), a maximum injection time of 3 ms and a normalized AGC target of 500% after fragmentation using higher-energy collisional dissociation (25% normalized collision energy).
Data processing
For identification, the data were searched against the Homo sapiens (UP000005640) UniProt database using Spectronaut (v.18.7 or v.19; Biognosys) by directDIA + analysis using default search settings. Enzyme specificity was set to trypsin, and a maximum of two missed cleavages were allowed. Carbamidomethyl was set as a fixed modification, and amino-terminal acetylation and oxidation of methionine were set as variable modifications. The resulting files were further processed using myProMS (v.3.10; https://github.com/bioinfo-pf-curie/myproms)54. For protein quantification, XICs from proteotypic peptides shared between compared conditions (TopN matching) with missed cleavages and carbamidomethylation were allowed. Median and scale normalization at the peptide level was applied on the total signal to correct the XICs for each biologically independent replicate (n = 5). To evaluate the significance of the change in protein abundance, a linear model (adjusted on peptides and biological replicates) was performed, and a two-sided t-test was applied on the fold change estimated by the model. The P values were then adjusted using the Benjamini–Hochberg false-discovery rate procedure. The MS proteomics raw data have been deposited to the ProteomeXchange Consortium through the PRIDE partner repository55.
Intranodal mouse metastasis models and in vivo administration of Fento-1
Mouse breast cancer cells (4T1) were transplanted into 6–8-week-old female Balb/c mice (syngeneic with the 4T1 model). Mice were housed at the Harvard Medical School animal core facility and randomized into separate cages. Mice were housed under sterile conditions with sterilized food and water provided ad libitum and maintained on a 12 h light–dark cycle and 22 ± 2 °C, 55 ± 5% humidity. To perform injections into lymph nodes, the lymphatics were first traced by injecting 2% Evans Blue dye (Sigma-Aldrich, E2129) into the foot pedal 5 min before performing intranodal injections. After injecting Evans Blue dye, the mice were anaesthetized using isoflurane and a small (5–10 mm) incision was made in the region of the right popliteal lymph node. The lymph node was located on the basis of Evans Blue staining, immobilized with forceps, and 1–2 × 104 cells suspended in 1× PBS were injected in a volume of 10 μl into the popliteal lymph node using a 27 G Hamilton syringe. Injection into the lymph node was confirmed by visible swelling of the lymph node. The incision was closed using surgical glue (3M VetBond Tissue Adhesive, 1469SB) and the mice were closely monitored for signs of pain or distress. Once tumours were palpable in at least 75% of the mice (approximately 1 week after injection), 10 μl Fento-1 (0.003 mg per animal) or vehicle was delivered intralymphatically into the tumour-bearing lymph node every other day until the experimental end point. Intranodal tumour diameters were measured 3 times per week with calipers until any tumour in the mouse cohort reached 2.0 cm in its largest diameter, which was the predetermined experimental end point for these experiments. For all experiments, this maximum permitted tumour diameter was not exceeded. At that point, all mice in the cohort were euthanized, per approved protocols, for analyses of intranodal tumour diameter, tumour mass and other parameters. At the experimental endpoint, data on tumour formation were collected in a manner blinded to sample identity or treatment. For survival-curve determinations, tumour-sized-based survival rate methods were applied, with 1.5 cm as the recommended predetermined survival rate per ethically acceptable cut-off values for determining therapeutic efficacy in mouse studies56. Tumour sizes were measured in two dimensions, length (x axis) and width (y axis), with the x axis conventionally defined as the longest axis of the tumour. Tumour sizes are represented as tumour diameter (cm) or tumour volume (cm3), as extrapolated by 0.5 × length × width2, as indicated for each in vivo experiment. Tumour samples were frozen in 10% DMSO in FBS (1 °C min–1 until –80 °C) for subsequent cellular analyses. No formal randomization techniques were used. However, animals were allocated randomly to treatment groups and specimens were processed in an arbitrary order. For all experiments, mice were kept on a normal chow diet and fed ad libitum.
Fluorescence imaging
Cells were plated on coverslips and treated as indicated. Bodipy 665/676 (Thermo Fisher Scientific, B3932, 10 µM, 45 min), LysoTracker Deep Red (Thermo Fisher Scientific, L12492, 100 nM, 45 min), DND-189 LysoSensor (Thermo Fisher Scientific, L7535, 100 nM, 1 h), Liperfluo (Dojindo, L248, 1 µM, 1 h) and SQSS (in-house, 50 nM, 1 h) were added to live cells before fixation. For colocalization studies, cells were treated with Fento-1 (1 µM, 1 h) or marmycin (in-house, 1 µM, 1 h) at the indicated temperature 16 h after transduction. At 16 h after transduction the BacMam-transduced cells were treated with RSL3 (1 µM) for 15 min or for 3 h 15 min and then treated with Bodipy 665/676 for 45 min to give the final time points of 1 h and 4 h. For colocalization of lipid peroxides with lysosomes, cells were treated with RSL3 (1 µM) and either Bodipy 665/676 and DND-189 LysoSensor or Liperfluo and LysoTracker Deep Red for 1 h. To assess the fluorescence intensity of Fento-1 in the presence of α-tocopherol, cells were pretreated with α-tocopherol (Sigma-Aldrich, PHR1031, 100 µM, 2 h) and then with Fento-1 (1 µM). Cells were then washed 3 times with 1× PBS, fixed with 2% paraformaldehyde in 1× PBS for 12 min and then washed 3 times with 1× PBS. For antibody staining, cells were permeabilized with 0.1% Triton X-100 in 1× PBS for 5 min and washed 3 times with 1× PBS. Subsequently, cells were blocked in 2% BSA (Euromedex, 04-100-812-E), 0.2% Tween-20 and 1× PBS (blocking buffer) for 20 min at room temperature. Cells were incubated with the relevant antibody in blocking buffer for 1 h at room temperature, washed 3 times with 1× PBS and were incubated with secondary antibodies for 1 h. Finally, coverslips were washed 3 times with 1× PBS and mounted using Vectashield containing DAPI (Vector Laboratories, H-1200-10). Bodipy 665/676 and Liperfluo-treated cells were fixed using ice-cold reagents and placed at 4 °C immediately after mounting on coverslips and imaged. Fluorescence images were acquired using a Deltavision real-time microscope (Applied Precision), a thunder microscope (Leica) or an Apotome Weiss microscope with Deltavision imaging software, Thunder Leica image acquisition software or Apotome Zeiss Imaging software, respectively. A ×40/1.4NA, ×60/1.4NA or ×100/1.4NA objective was used for acquisitions, and all images were acquired as z stacks. Images were deconvoluted using SoftWorx (ratio conservative, 15 iterations, Applied Precision) and processed with Fiji 2.0.0-rc-69/1.52n. Images were taken in black and white and colouring was applied with Fiji. Fluorescence intensity is displayed as arbitrary units and is not comparable between different panels. Colocalization quantification was calculated using Fiji 2.0.0-rc-69/1.52n. Nuclei were detected using DAPI or Hoechst fluorescence as indicated.
Bright-field microscopy and digital photographs
Bright-field images were acquired using a CKX41 microscope (Olympus) and cellSens Entry imaging software (Olympus).
Small-molecule labelling using click chemistry
In-cell click labelling
Cells on coverslips were treated as indicated with cDFO (in-house, 100 µM, 15 min), cLip-1 (in-house, 1 µM, 1 h or 2 h), metcLip-1 (in-house, 10 µM, 1 h), alcLip-1 (in-house, 10 µM, 1 h) or cCW (1 µM, 1 h), then fixed and permeabilized as indicated in the section ‘Fluorescence imaging’. LysoTracker Deep Red was added to live cells for 45 min before fixation. The click reaction cocktail was prepared using a Click-iT EdU Imaging kit (Invitrogen, C10337) according to the manufacturer’s protocol. In a typical experiment, we mixed 50 μl of 10× Click-iT reaction buffer with 20 μl CuSO4 solution, 1 μl Alexa-Fluor-azide, 50 μl reaction buffer additive (sodium ascorbate) and 379 µl ultrapure water to reach a final volume of 500 μl. Coverslips were incubated with the click reaction cocktail in the dark at room temperature for 30 min, then washed 3 times with 1× PBS. Immunofluorescence imaging was then performed as described in the section ‘Fluorescence imaging’.
Click labelling in tissues
Kidney and liver tissue samples collected from cLip-1-treated mice were fixed in 4% paraformaldehyde in 1× PBS overnight at 4 °C. Fixed tissues were incubated in 10% sucrose in 1× PBS for 30 min and then incubated in 20% sucrose in 1× PBS at 4 °C for 4 h, followed by embedding in OCT mounting compound (TissueTek, Sakura) on dry ice and stored at −80 °C. Frozen tissues were cut into 5-µm-thick sections using a Cryostat Microm HM 560 (Thermo Fisher Scientific) at −30 °C. Tissue sections were post-fixed with 1% paraformaldehyde in 1× PBS for 10 min and subsequently incubated with 100% acetone for 10 min at −20 °C. Sections were incubated in blocking solution (1× PBS containing 3% BSA and 0.2% Triton X-100) for 30 min. The specimens were labelled using a click reaction as described above. For costaining with a lysosomal marker, the click-labelled specimens were incubated with anti-LAMP1 or antibody diluted in 1× PBS containing 10% normal goat serum overnight at 4 °C. The next day, sections were incubated with secondary antibody in 1× PBS containing 1% BSA and 0.3% Triton X-100 for 2 h at room temperature. Nuclei were visualized with Hoechst 33342 staining, and slides were mounted in Aqua/PolyMount (Polysciences). Images were acquired using a fluorescence microscope (DS-Qi2, Nikon) or a confocal microscope (LSM880, Carl Zeiss) and the corresponding appropriate filter sets for fluorophores.
Western blotting
Cells were treated as indicated and then washed with 1× PBS. Proteins were solubilized in 2× Laemmli buffer containing benzonase (VWR, 70664-3, 1:100). Extracts were incubated at 37 °C for 1 h, heated at 94 °C for 10 min and quantified using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific). Protein lysates were resolved by SDS–PAGE electrophoresis (Invitrogen sure-lock system and Nu-PAGE 4-12% Bis-Tris precast gels). In a typical experiment, 10–20 µg of total protein extract were loaded per lane in 2× Laemmli buffer containing bromophenol blue. On each gel, a size marker was run: 3 µl PageRuler (Thermo Fisher Scientific, 26616) or 3 µl PageRuler plus (Thermo Fisher Scientific, 26620) and 17 µl 2× Laemmli buffer. Proteins were then transferred onto nitrocellulose membranes (Amersham Protran 0.45 μm) using a Trans-Blot SD semi-dry electrophoretic transfer cell (Bio-Rad) using 1× NuPage transfer buffer (Invitrogen, NP00061) with 10% methanol. Membranes were blocked with 5% non-fat skimmed milk powder (Régilait) in 0.1% Tween-20 and 1× PBS for 20 min. Membranes were cut at the appropriate marker size to enable the probing of several antibodies on the same membrane. Blots were then probed with the relevant primary antibodies in 5% BSA, 0.1% Tween-20 and 1× PBS or in 5% non-fat skimmed milk powder in 0.1% Tween-20 and 1× PBS at 4 °C overnight with gentle motion in a hand-sealed transparent plastic bag. Membranes were washed with 0.1% Tween-20 and 1× PBS 3 times and incubated with horseradish-peroxidase-conjugated secondary antibodies (Jackson Laboratories) in 5% non-fat skimmed milk powder (Régilait) in 0.1% Tween-20 and 1× PBS for 1 h at room temperature and washed 3 times with 0.1% Tween-20 and 1× PBS. Antigens were detected using SuperSignal West Pico PLUS (Thermo Fisher Scientific, 34580) and SuperSignal West Femto (Thermo Fisher Scientific, 34096) chemiluminescent detection kits. Signals were recorded using a Fusion Solo S Imaging System (Vilber) with FusionCapt Advance software. Images were processed using Fiji 2.0.0-rc-69/1.52n software. Full scans of blots are provided in the Supplementary Information.
Transduction
PDAC053T cells were seeded in 4-well plates at a density of 2 × 105 cells per well 24 h before the experiment in 1 ml cell culture medium. Cells were then transduced with CellLight Lysosomes-GFP, BacMam 2.0 (Thermo Fisher Scientific, C10596), CellLight ER-GFP, BacMam 2.0 (Thermo Fisher Scientific, C10590) or CellLight Mitochondria-GFP, BacMam 2.0 (Thermo Fisher Scientific, C10600) according to the manufacturer’s instructions. In brief, 70 µl BacMam reagent was added to the medium and mixed. Cells were incubated for 16 h.
RNA interference
Cells were plated 24 h before the experiments. Cells were transfected with the specified siRNA using jetPRIME (Polyplus, 114-15) according to the manufacturer’s protocol with 100 nM siRNA. In brief, PDAC053T cells were seeded in 6-well plates at a density of 2 × 105 cells per well and transfected 24 h later. The medium was replaced after 6 h. Analysis was performed 72 h after transfection. Suitable siRNAs were designed by Dharmacon for specific downregulation of target genes. siRNA sequences are provided in the Supplementary Information.
Flow cytometry
Cells were washed with ice-cold 1× PBS. For antibody staining, cells were incubated with Fc block (Human TruStain FcX, BioLegend, 422302, 1:20) for 15 min, then incubated with antibodies in ice-cold 10% FBS, 1× PBS, 2 mM EDTA for 20 min at 4 °C and then washed with 1× PBS and resuspended in 10% FBS, 1× PBS and 2 mM EDTA before analysis using a BD LSR Fortessa X-20 flow cytometer with FACS DIVA software (v.9.0.1).
Flow cytometry with iron probes
PDAC053T and HT-1080 cells were seeded in 6-well plates at a density of 2 × 105 cells per well. On the next day, Lip-1 (10 µM), hydroxychloroquine (Sigma-Aldrich, H0915, 100 µM) or bafilomycin A1 (Sigma-Aldrich, B1793, 75 nM) was added to PDAC053T cells for 1 h and to HT-1080 cells for 30 min. For PDAC053T cells, after 30 min of treatment with compounds, RPE27 (in-house, 40 µM) or HMRhoNox-M28 (in-house, 1 μM) probes were added for 30 min. For HT-1080 cells, after 15 min of treatment with compounds, RPE27 (in-house, 40 µM) or HMRhoNox-M28 (in-house, 1 μM) probes were added and left for 15 min. After incubation with the iron probes, the medium was removed and cells were washed with 1× PBS once before trypsinization. Cells were collected, pelleted, washed with 1× PBS and finally 200 µl of 1× PBS buffer containing 10% FBS and EDTA (0.1% v/v) was added. Data were recorded on a BD LSR Fortessa X-20.
Flow cytometry with Bodipy-C11 581/591
PDAC053T cells were seeded in 6-well plates at the density of 1 × 105 cells per well. On the next day, cells were treated with Lip-1 (1 µM), cLip-1 (in-house, 1 µM), metclip-1 (in-house, 10 µM), alcLip-1 (in-house, 10 µM) and Bodipy-C11 581/591 (200 nM) before adding RSL3 (100 nM). After 1 h, the medium was removed and cells were washed with 1× PBS twice before trypsinization. Cells were collected, pelleted, washed with 1× PBS and finally 250 µl of 1× PBS buffer containing 10% FBS and EDTA (0.1% v/v) was added for flow cytometry. PDAC053T, HT-1080 and 4T1 cells were seeded in 6-well plates at a density of 2 × 105 cells per well. On the next day, cells were treated with bafilomycin A1 (75 nM) and hydroxychloroquine (10 µM) for 2 h before adding RSL3 (200 nM for PDAC053T and HT-1080, 500 nM for 4T1). After 1 h, cells were treated with Bodipy-C11 581/591 (4 µM) for an additional 1 h. The medium was removed and cells were washed with 1× PBS twice before trypsinization. Cells were collected, pelleted, washed with 1× PBS and finally 250 µl of 1× PBS buffer containing 10% FBS and EDTA (0.1% v/v), was added for flow cytometry. Data were recorded on an AttuneTM NxT flow cytometer (Thermo Fisher Scientific) using Attune NxT (v.4.2.0).
Analyses of lysosomal glutathione and lysosomal hydroxy radicals
Cells were incubated with SQSS (in-house, 100 nM, 1 h) or 1-Red (in-house, 100 nM, 24 h). HT-1080 cells were incubated with RSL3 (1 µM), ML210 (10 µM, Sigma-Aldrich, SML0521), FIN56 (5 µM, Sigma-Aldrich, SML1740), buthionine sulfoximine (10 µM, Sigma-Aldrich, B2515) or erastin (10 µM, Sigma-Aldrich, 329600) for the indicated time points. Data were recorded on an AttuneTM NxT flow cytometer (Thermo Fisher Scientific).
Analyses of lysosomal iron content
Typically, 2 × 105 dissociated human tissue cells were incubated in medium (RPMI 1610, 10% FBS and penicillin–streptomycin) with HMRhoNox-M28 (in-house, 1 μM, 1 h). Lysosomal iron content of human tumour samples and healthy adjacent tissues was analysed with the following antibody and stain panel: DAPI (0.1 µg ml–1), CD3-BV510 (BioLegend, 317332), CD31-PE-Cy7 (BioLegend, 303118), CD44-AF647 (Novus Biologicals, NB500-481AF647), CD45-BV785 (BioLegend, 304048), CD163-PerCP/Cyanine5.5 (BioLegend, 326512), FAP-AF700 (R&D Systems, FAB3715N) and TFR1-APC-AF750 (Beckman Coulter, A89313). The live tumour cancer cells corresponded to DAPI–CD45–CD31–FAP– cells. Data were recorded on a BD LSRFortessa X-20 or AttuneTM NxT flow cytometer (Thermo Fisher Scientific).
Analyses of CD44 in Fento-1-treated tumour samples
Typically, 2 × 105 dissociated tumour cells were incubated in medium (RPMI 1610, 10% FBS and penicillin–streptomycin) with Fento-1 (in-house, 1 µM, 24 h). Cells were pretreated with Lip-1 (1 µM), cLip-1 (in-house, 1 µM), α-tocopherol (100 µM) or deferiprone (100 µM) for 2 h. α-Tocopherol was kept pure under inert atmosphere, and a fresh stock solution was prepared throughout the study before each experiment. The following antibody and stain panel was used for subsequent flow cytometry analysis: Sytox blue (Thermo Fisher Scientific, S34857, 1 µM), CD31-BV605 (BioLegend, 303122), CD45-BV510 (BioLegend, 368526, lot B373428), CD44-AF647 (Novus Biologicals, NB500-481AF647), TFR1-PE (BioLegend, 334106) and FAP-AF750 (Novus Biologicals, FAB3715S-100UG). The live tumour cells corresponded to Sytox blue–CD45–CD31–FAP– cells. Data were recorded on an AttuneTM NxT flow cytometer (Thermo Fisher Scientific). For flow cytometry analyses of CD44 levels in mouse 4T1 tumours, typically, 2 × 105 dissociated tumour cells were used per condition. Freshly dissociated cells were stained using the following antibody and stain panel: Sytox blue (Thermo Fisher Scientific, S34857, 1 µM), CD31-BV605 (BioLegend, 102427), CD44-AF647 (BioLegend, 103018), CD45-BV510 (BioLegend, 103138) and MHCII-APC/Cyanine7 (BioLegend, 107628). The live tumour cancer cells corresponded to Sytox blue–CD45–CD31–MCHII+ cells. Data were recorded on an AttuneTM NxT flow cytometer (Thermo Fisher Scientific). All data were analysed using FlowJo (v.10.10.0).
FACS
Sorting of human cells was performed using the following antibodies: CD31-PE-Cy7 (BioLegend, 303118), CD44-AF647 (Novus Biologicals, NB500-481AF647) and CD45-BV785 (BioLegend, 304048). The sorted cells corresponded to CD45–CD31–CD44+ cells and CD45–CD31–CD44– cells, which were isolated on a FACSAria Fusion (BD) using FACS DIVA (v.9.0.1). ICP-MS experiments were conducted using CD44high and CD44low tumour cells as described in the section ‘ICP-MS’. Sorting of mouse cells was performed using the following antibodies: CD44-AF647 (BioLegend, 103018) and MHCII-APC/Cyanine7 (BioLegend, 107628). The sorted cells corresponded to MHCII+CD44high cells and MHCII+CD44low cells. Sorted cells were centrifuged at 300g and cell pellets were processed for subsequent applications.
ICP-MS
Glass vials equipped with Teflon septa were cleaned with nitric acid 65% (VWR, Suprapur, 1.00441.0250), washed with ultrapure water (Sigma-Aldrich, 1012620500) and dried. Cells were collected and washed twice with 1× PBS. Cells were then counted using an automated cell counter (Entek) and transferred in 200 µl 1× PBS or ultrapure water to the cleaned glass vials. The same volume of 1× PBS or ultrapure water was transferred into separate vials for background subtraction, at least in duplicate per experiment. For tissue samples, a small piece of about 1 mm3 was transferred into a clean preweighed vial. Samples were lyophilized using a freeze dryer (Christ, 2-4 LDplus). Vials with tissue samples were subsequently weighed to determine the tissue dry weight. Samples were then mixed with nitric acid 65% and heated at 80 °C overnight in the same glass vials closed with a lid carrying a Teflon septum. Samples were then cooled to room temperature and diluted with ultrapure water to a final concentration of 0.475 N nitric acid and transferred to metal-free centrifuge vials (VWR, 89049-172) for subsequent MS analyses. Amounts of metal were measured using an Agilent 7900 ICP-QMS in low-resolution mode, taking natural isotope distribution into account. Sample introduction was achieved with a micronebulizer (MicroMist, 0.2 ml min–1) through a Scott spray chamber. Isotopes were measured using a collision–reaction interface with helium gas (5 ml min–1) to remove polyatomic interferences. Scandium and indium internal standards were injected after inline mixing with the samples to control the absence of signal drift and matrix effects. A mix of certified standards was measured at concentrations spanning those of the samples to convert count measurements to concentrations in the solution. Values were normalized against cell number or tissue dry weight.
Liposome preparation and lipid oxidation studies
Liposomal structures were prepared using the traditional lipid film hydration method. In brief, 100 μl of a stock solution (1 mg ml–1 chloroform) of 18:1 (Δ9-cis) PC (DOPC, Avanti Polar Lipids) was dissolved in 400 μl chloroform and transferred into a round-bottom flask. The organic solvent was removed under reduced pressure in a rotary evaporator for 15 min at 200 r.p.m. in a water bath at 37 °C. Afterwards, the lipid film was dried with a vacuum pump overnight. The sample was hydrated with 1 ml of 0.1 mM sodium acetate buffer (pH 4.5) and vortexed every 5 min for 20 min. Liposomes were extruded by passing the suspension through 2 polycarbonate membranes (pore size 0.2 mm) 20 times. For the control experiment, 200 μl of the liposome solution was added into an Eppendorf tube and heated at 37 °C with agitation at 800 r.p.m. Then, 5 μl of an aqueous solution of iron(II) triflate (1.4 mg in 1.5 ml) and 13 μl of 0.1 mM acetate buffer (pH 4.5) were added. At t = 0 min 13 μl of an aqueous solution of H2O2 (10 μl H2O2 (30%) in 1 ml) was added. For the Fento-1 experiment, 200 μl of the liposome solution was added to an Eppendorf tube and heated at 37 °C at 800 r.p.m. Then, 13 μl of a 1 mM solution of Fento-1 in DMSO and 5 μl of an aqueous solution of iron(II) triflate (1.4 mg in 1.5 ml) were added. At t = 0 min, 13 μl of an aqueous solution of H2O2 (10 μl H2O2 (30%) in 1 ml) was added. DOPC oxidation was recorded with a QExactive mass spectrometer (Thermo Fisher Scientific) equipped with a TriVersa NanoMate ion source (Advion Biosciences) as detailed in the section ‘MS-based lipidomics’. Samples were injected at 0.5 h, 1 h, 2 h, 3 h, 4 h, 7 h and 24 h reaction times.
Isolation of lysosome-enriched fractions
HT-1080 cells were treated with Fento-1 (10 µM) for 1 h, and lysosome-enriched fractions were isolated using a Lysosome Isolation kit (Abcam, ab234047) according to the manufacturer’s protocol. In brief, 2 × 107 cells were washed and centrifuged at 600g for 10 min and the supernatant was removed. Cells were resuspended in lysosome isolation buffer, vortexed and incubated on ice for 2 min. Complete cell disruption was obtained using a dounce homogenizer. After adding lysosome enrichment buffer, the homogenate was centrifuged at 500g for 10 min at 4 °C. The supernatant was added to the top of a discontinuous gradient density and an ultracentrifugation at 145,000g for 2 h at 4 °C was performed. The lysosome-enriched fraction was present in the top 10% of the gradient volume. For MS-based lipidomics, the lysosomal fraction was mixed with 2 volumes of 1× PBS, vortexed and centrifuged at 18,000g for 30 min at 4 °C. Next, 200 µl of 150 mM sodium bicarbonate was added to the pellet and the sample was flash-frozen in liquid nitrogen. Lipidomic analyses were performed on these flash-frozen samples. For western blot analyses, the protein content of the lysosomal-enriched gradient supernatant was quantified using a Qbit 1 fluorometer (Thermo Fisher Scientific) and a protein quantification kit (Thermo Fisher Scientific, Q33212). Equal total protein amounts of total cell extracts and lysosome-enriched extracts were loaded for comparison for western blot analyses.
MS-based lipidomics
For comparison of ferroptosis inducers, HT-1080 cells were treated with Fento-1 (in house, 1 µM), erastin (10 µM), RSL3 (100 nM) or iFSP1 (10 µM, Sigma-Aldrich, SML2749) for 24 h. For cotreatment with ferroptosis inhibitors, HT-1080 cells were pretreated with α-tocopherol (100 µM), deferiprone (100 µM) or Lip-1 (10 µM) for 2 h, and then with Fento-1 (1 µM) for 24 h. Dissociated human tumour samples were pretreated with 100 µM α-tocopherol and then with 1 µM Fento-1 for 24 h. PDAC053T cells were treated with 1 µM Fento-1 for 6 h or 24 h. Dissociated mouse tumour samples were counted and processed directly after dissociation and pretreated with α-tocopherol (100 µM) for 2 h and then with Fento-1 (1 µM, 24 h). Cells were subsequently washed with 1× PBS and then with 150 mM ammonium bicarbonate. Cells were then resuspended in 150 mM ammonium bicarbonate and centrifuged at 300g for 5 min. The supernatant was removed and cells were resuspended in 1 ml of 150 mM ammonium bicarbonate. The solutions were centrifuged at 12,000 r.p.m. for 10 min and the supernatant was removed. Next, 200 µl of 150 mM sodium bicarbonate was added to the pellet and samples were flash-frozen in liquid nitrogen. Lipidomic analyses were performed on the same day for all technical and biological replicates for a given dataset. For lipidomic analyses, 200 µl cell lysate was spiked with 1.64 μl internal standard lipid mixture containing 300 pmol phosphatidylcholine (PC) 17:0-17:0, 50 pmol phosphatidylethanolamine (PE) 17:0-17:0, 50 pmol phosphatidylinositol (PI) 16:0-16:0, 50 pmol phosphatidylserine (PS) 17:0-17:0, 30 pmol phosphatidic acid (PA) 17:0-17:0, 30 pmol phosphatidylglycerol (PG), 30 pmol lysophosphatidylcholine (LPC) 12:0, 30 pmol lysophosphatidylethanolamine (LPE) 17:1, 30 pmol lysophosphatidylserine (LPS) 17:1 and 30 pmol lysophosphatidic acid (LPA) 17:0 and subjected to lipid extraction at 4 °C as previously described57. In brief, the sample was extracted with 1 ml chloroform and methanol (10:1) for 2 h at 4 °C with vigorous shaking in a thermomixer (1,000 r.p.m.). The lower organic phase was collected and dried in a SpeedVac vacuum concentrator. The remaining aqueous phase was re-extracted with 1 ml chloroform and methanol (2:1) for 1 h at the same temperature and shaking conditions. The lower organic phase was collected and evaporated in a SpeedVac vacuum concentrator. Lipid extracts were dissolved in 100 μl infusion mixture consisting of 7.5 mM ammonium acetate dissolved in propanol, chloroform and methanol (4:1:2 (v/v/v)). For the in vitro liposome experiments, 1 µl of each reaction mixture taken at different time points was added to 100 μl infusion mixture consisting of 7.5 mM ammonium acetate dissolved in propanol, chloroform and methanol (4:1:2 (v/v/v)) containing 300 pmol PC 17:0-17:0. Samples were analysed by direct infusion in a QExactive mass spectrometer (Thermo Fisher Scientific) equipped with a TriVersa NanoMate ion source (Advion Biosciences). In brief, 5 µl of sample was infused with the gas pressure and voltage set to 1.25 psi and 0.95 kV, respectively. PC and oxidized PC (PCOx) were detected in the 10:1 extract by positive-ion mode FTMS as protonated adducts by scanning m/z = 580–1,000 Da, at Rm/z = 200 = 280,000 with lock mass activated at a common background (m/z = 680.48022) for 30 s. Every scan is the average of 2 microscans, and the AGC was set to 106 and the maximum ion injection time (IT) was set to 50 ms. PE, oxidized PE (PEOx) and LPE were detected as deprotonated adducts and LPC were detected as acetate adducts in the 10:1 extract by negative-ion mode FTMS by scanning m/z = 420–1,050 Da, at Rm/z = 200 = 280,000 with lock mass activated at a common background (m/z = 529.46262) for 30 s. Every scan is the average of 2 microscans, AGC was set to 106 and the maximum ion IT was set to 50 ms. PI, oxidized PI (PIOx), PS, oxidized PS (PSOx), lysophosphatidylinositol (LPI) and lysophosphatidylserine (LPS) were detected in the 2:1 extract, by negative-ion mode FTMS as deprotonated ions by scanning m/z = 400–1,100 Da, at Rm/z = 200 = 280,000 with lock mass activated at a common background (m/z = 529.46262) for 30 s. Every scan is the average of 2 microscans, the AGC was set to 106 and the maximum ion IT was set to 50 ms. Annotation of the oxidized phospholipids only details the number of additional oxygen atoms and double bonds present in a given mass and it can potentially refer to several different structures with the same monoisotopic mass. All data were acquired in centroid mode. All lipidomic data were analysed with the lipid identification software LipidXplorer (v.1.2.8; http://genomebiology.com/2011/12/1/R8). Tolerance for MS and identification was set to 2 ppm. Data were normalized against internal standards and total input to the respective lipid species. Lipidomic data were clustered using a correlation distance and Ward linkage, and represented as heat maps of normalized values.
Glycerol quantification
HT-1080 cells were treated with Fento-1 (in-house, 1 or 2 µM, 24 h). Glycerol was quantified using a Glycerol-glo assay (Promega, J3150) according to the manufacturer’s protocol. In brief, the assay was performed in a 96-well plate and 4,000 cells were plated per well 24 h before the experiment. A standard curve was prepared for each biological experiment and three technical replicates were performed for each condition and each biological replicate. Luminescence signals were recorded using a SpectraMax ID3 plate reader (Molecular Devices). Data were exported and analysed using Excel (Microsoft) and Prism software.
Software for illustrations
Illustrations were created using Fiji 2.0.0-rc-69/1.52n, Prism 10.0.3 and Adobe Illustrator 26.0.2. Illustrations in Figs. 2b and 4 were created using BioRender (https://biorender.com).
Statistical and reproducibility
Results are presented as the mean ± s.e.m. or s.d. as indicated. For box plots, boxes represent the interquartile range and median, and whiskers indicate the minimum and maximum values. Prism (v.10.0.3) software was used to calculate P values using a two-sided Mann–Whitney test, two-sided unpaired t-test, Kruskal–Wallis test with Dunn’s post-test, one-way ANOVA, two-way ANOVA or Mantel–Cox log-rank test as indicated. Prism (v.10.0.3) software was used to generate graphical representations of quantifications unless stated otherwise. Sample sizes (n) are indicated in the figure legends and were not predetermined. A minimum of n = 3 independent experiments were performed as a standard, and sample sizes were increased in more complex experiments to ensure reproducibility. Exact P values are indicated in the figures.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.