Data generation by institute
Metadata about scRNA-seq and CITE-seq samples, including information on source study, cell enrichment and donor age, are provided in Supplementary Table 1. Information about spatial data, including Visium, IBEX, RareCyte and RNAscope, is provided in Supplementary Table 2. In brief, all CITE-seq data were generated at Ghent University and mapped to the human genome (GRCh38) at the Wellcome Sanger Institute (WSI). All other original scRNA-seq data and 10x Visium data were generated and mapped to the human genome (GRCh38) at WSI. All IBEX imaging was performed at the National Institute of Allergy and Infectious Diseases (NIAID), NIH. All non-IBEX imaging datasets (RareCyte, RNAscope, Visium H&E) were generated at WSI. No fetal work was performed at the NIH and at Ghent University.
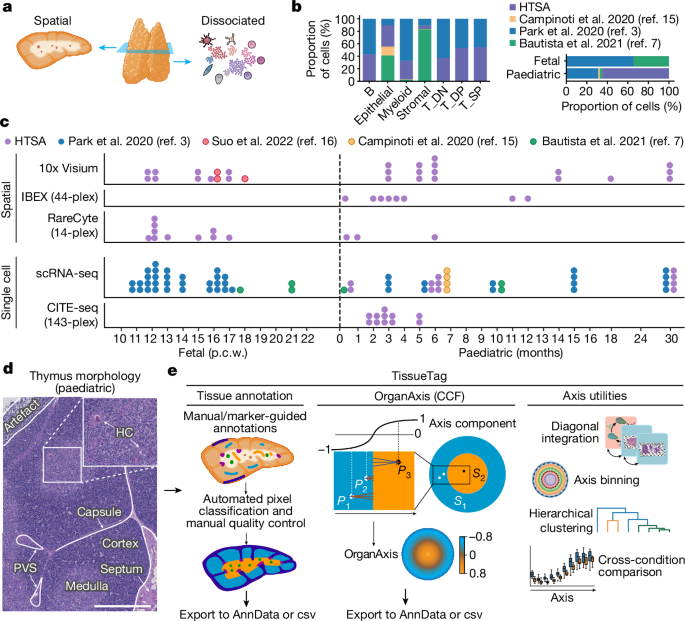
Human fetal and paediatric data from several previous studies3,7,15,16 were included and reanalysed from raw fastq files. Details on sample processing, ethics and funding are available in the respective publications; details on the origin of each sample are provided in Fig. 1c and Supplementary Tables 1 and 2.
For samples processed at WSI, paediatric samples were obtained from cardiac corrective surgeries and provided by Newcastle University collected under REC approved study 18/EM/0314 and Great Ormond Street Hospital under REC approved study 07/Q0508/43. Human embryonic and fetal material was provided by the joint MRC & Wellcome Trust (grant MR/006237/1) Human Developmental Biology Resource (http://www.hdbr.org) with written consent and approval from the Newcastle and North Tyneside NHS Health Authority Joint Ethics Committee (08/H0906/21+5). Samples processed at Ghent University were obtained according to and used with the approval of the Medical Ethical Commission of Ghent University Hospital, Belgium (EC/2019-0826) through the haematopoietic cell biobank (EC-Bio/1-2018). Samples processed at the NIH were obtained under NIAID MTA 2016-250 from the pathology department of the Childrenâs National Medical Center in Washington, DC, following cardiothoracic surgery from children with congenital heart disease. Use of these thymus samples for this study was determined to be exempt from review by the NIH Institutional Review Board in accordance with the guidelines issued by the Office of Human Research Protections. Informed consent was obtained from all donors or their legal guardians.
Sample processing and library preparation for 10x scRNA-seq at WSI
Surgically removed paediatric thymi were directly moved to HypoThermosol (Sigma-Aldrich, H4416-100ML), shipped by courier with ice packs, and processed within 24âh from time of surgery. For scRNA-seq experiments, we performed single-cell dissociation as described in the protocol available online (https://doi.org/10.17504/protocols.io.bx8sprwe). In brief, thymic tissue was finely minced and cell dissociation was performed using a mixture of liberase TH (Roche, 05401135001) and DNase I (Roche, 4716728001) for ~30â60âmin in two rounds. Digested tissue was filtered through a 70âμm strainer and digestion was stopped with 2% FBS in RPMI medium. Red blood cell lysis was then performed on the cell pellet using RBC lysis buffer (eBioscience, 00-4333-57), after which cells were washed and counted. Magnetic and/or FACS sorting were performed to enrich stromal populations. Magnetic sorting to enrich for EPCAM+ or to deplete CD45+ or CD3+ cells was performed for U09, U48 and Z11 samples (Supplementary Table 1) using a magnetic sorting kit from Miltenyi Biotec including LS Columns (130-042-401) and the following bead-tagged antibodies: human CD45 MicroBeads (130-045-801), human CD326 (EPCAM) MicroBeads (130-061-101), human CD3 MicroBeads (130-050-101). Z11 and U40 samples were FACS-sorted to enrich for CD45â stromal cells (Z11) or to obtain both CD45â cells and total TECs (U40) (Supplementary Table 1). To perform FACS sorting, cells were resuspended in FACS buffer (0.5% FBS and 2âmM EDTA in PBS), blocked with TruStainFcX (BioLegend, 422302) for 10âmin and were stained with antibodies against EPCAM (anti-CD326 PE, 9C4, BioLegend, 324206), CD45 (anti-CD45 BV785, HI30, BioLegend, 304048), CD205 (anti-CD205 (DEC-205) APC, BioLegend, 342207) and CD3 (anti-CD3 FITC, OKT3, BioLegend, 317306) and DAPI for 30âmin. All antibodies were diluted 1:50 for staining. After staining, cells were washed and sorted using Sony SH800 or Sony MA900 sorters with a 130âμm nozzle. Samples were first gated to remove debris and dead cells, but no singlet gate was applied to ensure large TECs would not be excluded. For samples stained with anti-CD45, total CD45â cells were sorted to enrich for stroma. For EPCAM/CD205-stained cells, EPCAM+CD205â and EPCAMâCD205+ cells were sorted to obtain the total TEC fraction including cortical and medullary epithelial cells but exclude autofluorescent cells (Supplementary Fig. 19a). FCS Express v.7.18.0025 was used for data analysis. After sorting/enrichment, cells were resuspended in collection buffer to the recommended concentration (106 cells per ml) and loaded to capture around 8,000â10,000 cells on the 10x Genomics chromium controller to generate an emulsion of cells in droplets using Chromium Next GEM Single Cell 5â² Kit v2 (1000263). GEX and TCR-seq libraries were further prepared using the Library Construction Kit (1000190) and Chromium Single Cell Human TCR Amplification Kit (1000252) according to the manufacturerâs instructions. Sequencing was performed on the NovaSeq 6000 sequencer (Illumina). Additional details are provided in Supplementary Table 1.
Curation of published datasets
Data from several previous studies3,7,15,16 are included in this Article. For public datasets deposited on ArrayExpress, paired-end fastq files were downloaded from ENA, and the .sdhf file was used to determine the type of experiment (3â²/5â² and the version of 10x Genomics kit). For public datasets deposited on GEO, if the data were deposited as a Cell Ranger .bam file, URLs for the bam files were obtained using srapath v.2.11.0. The .bam files were downloaded and converted to .fastq files using 10x bamtofastq v.1.3.2. If GEO data were deposited as paired-end fastq files, sra files were located using the search utility from NCBI entrez-direct v.15.6, downloaded and converted to fastq files using fastq-dump v.2.11.0. Sample metadata were curated from the abstracts deposited on GEO. Finally, published datasets that had been generated at WSI were downloaded from iRODs v.4.2.7 in the form of cram files and converted to fastq files using samtools v.1.12. using the command âsamtools collate -O -u -@16 $CRAM $TAG.tmp | samtools fastq -N -F 0Ã900 -@16 â1 $TAG.R1.fastq.gz â2 $TAG.R2.fastq.gz -â. Sample metadata were obtained using the imeta command from iRODs.
Processing of published and newly generated scRNA-seq and TCR-seq datasets at WSI
After fastq file generation, 10x Genomics scRNA-seq experiments were processed using the STARsolo pipeline detailed on GitHub (https://github.com/cellgeni/STARsolo). A STAR human genome reference matching Cell Ranger GRCh38-2020-A was prepared as per instructions from 10x Genomics. Using STAR v.2.7.9a and the previously collected data about sample type (3â²/5â², 10x Genomics kit version), we applied the STARsolo command to specify UMI collapsing, barcode collapsing, and read clipping algorithms to generate results maximally similar to the default parameters of the âcellranger countâ command in Cell Ranger v.6: â–soloUMIdedup 1MM_CR –soloCBmatchWLtype 1MM_multi_Nbase_pseudocounts –soloUMIfiltering MultiGeneUMI_CR –clipAdapterType CellRanger4 –outFilterScoreMin 30â. For cell filtering, the EmptyDrops algorithm of Cell Ranger v.4 and above was invoked using â–soloCellFilter EmptyDrops_CRâ. The option â–soloFeatures Gene GeneFull Velocytoâ was used to generate both exon-only and full length (pre-mRNA) gene counts, as well as RNA velocity output matrices. TCR-seq samples were processed using Cell Ranger v.6.1.1 with VDJ reference vdj-GRCh38-alts-5.0.0. The default settings of the reference-based âcellranger vdjâ command were used. Fastq files were converted to <Sample>_S1_L001_R1_001.fastq.gz format to be compatible with Cell Ranger.
scRNA-seq quality control, data integration and annotation
Jupyter notebooks used for data quality control, preprocessing, integration and annotations are available in the GitHub repository for this manuscript (Code availability). Scanpy v.1.9.1 with anndata v.0.10.7 and the statistics and plotting libraries pandas v.2.2.2, numpy v.1.26.4, scipy v.1.13.0, seaborn v.0.13.2 and matplotlib v.3.8.4 were used for data analysis and visualization. Mapped libraries were subjected to computational removal of ambient RNA using CellBender42 v.0.1.0. Next, all datasets underwent cell quality-control filtering and cells with <400 or > 6500 genes, >6% mitochondrial reads or <5% ribosomal counts were removed. Doublets were annotated using Scrublet43 v.0.2.3. Next, datasets were integrated with scVI from scvi-tools44 v.0.19.0, for which mitochondrial, TCR and cell cycle genes were removed, and cells were annotated into major lineages (cell_type_level_0: T_DN, T_DP, T_SP, Epithelial, Stroma, Myeloid, RBC, B and Schwann) by Leiden clustering. Individual cell lineages were then separated and integrated with scVI to perform fine-grained annotation and remove remaining doublets picked up by manual annotation. Cell annotations were assigned based on four sequential steps: (1) high-resolution Leiden clustering was performed to find all potential cell clusters. (2) Annotations of new cells generated in this study were predicted (i) based on KNN graph majority voting of neighbouring cells with annotations from previous studies by adaptation of the weighted KNN transfer solution from scArches45, or (ii) automatic label transfer using CellTypist46 v.1.6.2 with the Developing_Human_Thymus or Pan_Fetal_Human models. (3) Calling the cell type annotation for a given cluster was then informed by a combination of the predicted labels and by marker genes reported in the literature. (4) Additional QC was performed and newly detected doublet clusters were removed where applicable. Steps 1â4 were repeated until final, fine-grained annotations were reached for the highest resolution (cell_type_level_4). Importantly, cell clusters that passed quality control but that we could not confidently assign to a defined cell type either in the literature or by cell markers by the strategy described above, were kept in the integrated object as âcell_type_level_4_exploreâ, which we recommend for future exploration and validation of these cell states. Annotations in âcell_type_level_4â were grouped into five hierarchical levels from the finest (cell_type_level_4) to the broadest (cell_type_level_0) (Supplementary Table 7).
Single-cell TCR-sequencing data were processed using Dandelion47 v.0.3.1, and a detailed notebook can be found in the GitHub repository for this Article (see Code availability). In brief, a Dandelion class object with n_obs was constructed from all combined TCR libraries. Cells were then further subset to only include DP and SP subtypes that contained V and J rearrangements for both TRB and TRA loci. Next, milo neighbourhoods were constructed based on scVI neighbourhood graphs (n_neighbors = 100) and VDJ genes and frequency feature space was calculated.
mTEC trajectory analysis
The scFates package48 v.1.0.7 was used for trajectory analysis on the combined mTECII and mTECIII cells. mTECII and mTECIII cells from the paediatric scRNA-seq dataset were reintegrated and batch-corrected using scVI. Next, the object was preprocessed according to recommendations from Palantir49 and the scVI latent embedding was used as an input to âpalantir.utils.run_diffusion_mapsâ (Palantir v.1.3.3). Tree learning was performed using scf.tl.tree by using multiscale diffusion space from Palantir as recommended by scFates. Next, the node that was characterized by expression of some mTECI markers (ASCL1, CCL21) together with mTECII markers (AIRE, FEZF2) was used as a root to compute pseudotime using scf.tl.pseudotime. Finally, milestones obtained after running the pseudotime were used to adjust the annotations. To derive differentially expressed genes across pseudotime branches, we applied the scf.tl.test_fork function, followed by scf.tl.branch-specific as described in the scFates article48. In brief, it fits a generalized additive model for each gene using pseudotime, branch and interaction between pseudotime and branch as covariates; two-sided P values were extracted for the interaction term (pseudotime:branch) and corrected using FDR to obtain significant differentially expressed genes. Next, these genes were tested for upregulation in each branch and assigned to different branches based on a cut-off of 1.3-fold upregulation.
mcTEC differentiation potential analysis
The STEMNET package50 (v.0.1) was used to determine the differentiation potential of mcTEC progenitor cells. For this purpose, only mTEC, cTEC and mcTEC(-Prolif) cells were retained in the dataset. mTECI/II/III and cTECI/II/III were set as maturation endpoints and the probability of each cell to adopt any of these six possible fates was calculated. To identify priming of mcTEC(-Prolif) towards mTECI versus cTECI fate, we derived a priming score by calculating the difference between the posterior probabilities for these two fates for each cell. Cells with priming score <â0.5 or >0.5 were labelled as mTECI-primed and cTECI-primed, respectively, whereas all other cells with comparable mTECI and cTECI potential were deemed to be unprimed.
Formulation of the CMA with OrganAxis
OrganAxis is a mathematical model aimed to derive the relative, signed position of a point in space in respect to two morphological landmarks. The OrganAxis base function H is highly flexible and tuneable with respect to the research question, spatial resolution and sampling frequency. In Supplementary Notes 1 and 2, we provide a detailed guide on tissue annotations with TissueTag v.0.1.1 and the details of OrganAxis derivation. The CMA is an extrapolation of OrganAxis, which is defined by a weighted linear combination of two H functions (CMAâ=â0.2âÃâH(edge-to-cortex) + 0.8âÃâH(cortex-to-medulla)). All IBEX and Visium images were annotated with TissueTag v.0.1.1 at a resolution of 2 um per pixel (ppmâ=â0.5). Then, annotations were transferred to a quasi-hexagonal grid that was generated by placing points with r-microns spacing in the x and y directions and staggering every other row by r/2. Throughout this study we used râ=â15. L2 distances to broad level annotations (annotation_level_0) and the corresponding CMA values were calculated with kâ=â10 for all spots in the hexagonal grid. L2 distances to fine annotations (annotation_level_1) were calculated with k = 1. CMA and L2 distances were then transferred to spots in Visium datasets or nuclei segmentations for IBEX by nearest-neighbour mapping and were therefore spatially homogeneous for these two spatial technologies.
To provide a common reference, we also binned the axis to a sequential discrete space for the entire thymus by ten levels (capsular, subcapsular, cortical level 1, cortical level 2, cortical level 3, cortical CMJ, medullary CMJ, medullary level 1, medullary level 2, medullary level 3; details are provided in Supplementary Note 2 and Supplementary Table 8).
10x Visium spatial transcriptomics sample processing and sequencing
Resected fetal and paediatric thymi were directly moved to HypoThermosol (Sigma-Aldrich, H4416-100ML) (paediatric samples) or cold PBS (fetal samples), shipped by courier with ice packs and processed within 24âh from time of surgery. For embedding, fetal or paediatric thymus tissue was first transferred to PBS and then placed onto ice for a few minutes to clear away any excess medium and preservation liquid (such as HypoThermosol). Next, as much liquid as possible was removed from the sample and, if necessary, the tissue was trimmed to fit into a cryomold. The sample was placed into a cryomold (Tissue-Tek, AGG4581) filled with OCT (Leica biosystems, 14020108926) and positioned according to the desired orientation. The cryomold was then placed in isopentane that had been equilibrated to â60â°C and left to fully freeze for 2âmin. The sample was then rested on dry ice to allow draining of the isopentane. Finally, the cryomolds were wrapped in foil and stored at â80â°C. On the day of sectioning, the samples were removed 1âh before sectioning and placed into the cryostat (Leica biosystems) at â18â°C to equilibrate. Tissue was sectioned (section thicknesses are provided in Supplementary Table 2) and sections were placed onto Visium slides according to the manufacturerâs protocol. The sections were stained with H&E and imaged at Ã20 magnification (Hamamatsu Nanozoomer 2.0 HT). Libraries were further processed according to the manufacturerâs protocol (Visium Spatial Gene Expression Slide & Reagent Kit, 10x Genomics, PN-1000184) (permeabilization times are shown in Supplementary Table 2). The samples were sequenced on the NovaSeq 6000 sequencer (Illumina) and the obtained fastq files were mapped with Space Ranger (10x Genomics; version numbers are provided in Supplementary Table 2).
Visium preprocessing, image registration and annotation
To process the Visium histology image data in higher resolution than the SpaceRanger defaults, we built a custom pipeline to extract an additional layer of image resolution at up to 5,000 pixels (hires5K), which we found to be more suitable for morphological analysis. We also developed our own fiducial image registration pipeline for increased accuracy where the fiducials are detected with cellpose v.2.1.1 and RANSAC from scikit-image v.0.22.0 is used for affine registration of reference fiducial frame (information provided by 10x Genomics). Lastly, for flexible tissue detection, we used Otsu thresholding with an adjustable threshold.
We subsequently used TissueTag v.0.1.1 for semiautomated image annotation (Supplementary Note 1). Cortical and medullary pixels were predicted with a pixel random forest classifier by generating training annotations based on spots with the highest gene expression of AIRE (for medulla) and ARPP21 (for cortex). Automatic cortex/medulla annotations were then adjusted manually where necessary. Moreover, we manually annotated individual thymic lobes and specific structures, such as capsule/edge, freezing/sectioning artefacts, HCs, PVS and fetal thymus-associated lymphoid aggregates (as defined previously16). The morphological annotation and evaluations were done in consultation with expert human thymic pathologists. A full example of the Visium processing pipeline and annotation is provided on the GitHub repository for this Article (see Code availability).
Spatial Visium mapping with cell2location
To ensure the best possible matching between Visium and single-cell profiles, we performed spatial mapping using cell2location51 v.0.1.3 separately for fetal and paediatric datasets. We therefore subset our single-cell reference datasets according to either fetal or paediatric stage and removed rare cell types that were predominantly found in one of these stages. We then further removed cell types that showed stress signatures (which we believed to originate from technical factors) and cell types with the total number of cells < 40. A list of cell types that were excluded and the exclusion criteria are provided in Supplementary Table 9. Before cell2location deconvolution, we removed cell cycle genes (Supplementary Table 10) and mitochondrial genes, as well as TCR genes using the regex expression â^TR[AB][VDJ]|^IG[HKL][VDJC]â. We then further calculated highly variable genes and used relevant metadata cofactors (sample, chemistry, study, donor, age) to correct for batch effects in the cell2location model.
Quality control and batch correction of Visium data
After deconvolution, all Visium data were subjected to filtering based on read coverage and predicted cell abundance. Spots with fewer than 1,000 genes per spot or fewer than 25 predicted cells were omitted. Furthermore, annotated tissue artefacts and areas not assigned to a specific structure were removed. Next, to generate a common embedding we performed scVI integration after removing cell cycle, mitochondrial and TCR genes from the highly variable gene selection. scVI training was performed with âSampleIDâ as the batch key, âSlideID, Spaceranger, section_thickness(um)â as categorical covariates, and âAge(numeric), n_genes_by_countsâ as continuous covariates.
Before performing any association analysis with the CMA, we further removed lobules (based on âannotations_lobules_0â) that had no or small medullar or cortical  regions, as we expected our CMA model to be less accurate in these cases.
PCA cumulative contribution of CMA
To estimate the dependency of the axis on spot gene variance across samples, we first normalized to a target sum of 2,500 counts and performed log transformation followed by combat regression (https://scanpy.readthedocs.io/en/stable/api/generated/scanpy.pp.combat.html) by sample to adjust for the batch effect of individual samples. We then computed the PCA for batch-corrected gene expression and calculated the Spearman correlation between the first ten PCs and CMA or the number of genes detected per spot (n_gene_by_counts). Note that number of genes per spot, in our hands, was mostly influenced by inconsistent permeabilization during Visium library preparation and constituted the largest technical source of within-sample variance that we found in both fetal and paediatric Visium samples. To estimate the cumulative contribution of either CMA or the number of genes per spot, we multiplied the Spearmanâs R with the percentage cumulative explained variance of the first 10 PCs.
Cytokine clustering by expression along the binned axis in Visium data
To analyse cytokine gradients based on the spatial distribution across CMA bins, we first selected a group of 65 cytokines that were broadly expressed from the CellphoneDB database (v.4.1.0, genes annotated as âCytokineâ, âgrowthfactor | cytokineâ, âcytokineâ, âcytokine | hormoneâ). We excluded cytokines that were expressed in less than 5% of the spots in all CMA bins. We then performed hierarchical clustering on gene expression batch-corrected (combat) fetal Visium samples of the standardized mean expression of genes across bins using the Ward linkage method with the linkage function from the scipy.cluster.hierarchy module. A heat map was generated using the matrixplot function from the scanpy package52.
Cytokine two-way ANOVA analysis and cosine similarity on binned axis Visium data
To compare the distribution of cytokines across developmental groups (fetal versus paediatric) and identify differentially distributed genes, we implemented a two-way ANOVA approach. We initially log-normalized Visium gene expression, then removed lobules for which not a single cortex or medulla Visium spot was detected to increase CMA confidence in both datasets. Data were grouped by mean expression per sample and CMA bin, such that each sample had a single datapoint per CMA bin; nâ=â16 (paediatric) and nâ=â12 (fetal) samples. Cosine similarity was calculated based on the median values of the pooled sample bins between fetal and paediatric gene profiles with sklearn.metrics.pairwise.cosine_similarity from scikit-learn v.0.22.0. Two-way ANOVA for age group (fetal versus paediatric) and CMA bin was calculated with statsmodels.api.stats.anova_lm(model, type=2). P values for main and interaction effects were Bonferroni corrected with statsmodels.stats.multitest.multipletests (pvals, alpha=0.05, method=âbonferroniâ). For the full report of the results refer to Supplementary Table 5.
Cell- and gene-weighted mean CMA location on Visium data
To estimate the average position of a cell or gene distribution along CMA and HC axes (L2 distance to the nearest HC), spots with low gene expression were filtered out by using appropriate thresholds (0.2 for scVI corrected gene expression and 0.5 for predicted cell abundances). The position of a gene or cell was then calculated according to the following formula: for every gene/cell and axis positions, the weighted mean was calculated as a dot product of spot cell abundance values and CMA position divided by the sum of the cell abundance values across spots.
Identification of cell-type-specific SGs
To identify genes exclusively expressed in a specific cell type or subset thereof (âspecialization genesâ, SGs), we developed custom Python functions. Starting from raw read count, gene expression was scaled with scipy.stats.zscore(). Cells that showed expression below a cut-off of 0.05 and genes that had expression below 1.5 mean counts were excluded from further steps. Next, a quantile threshold (>95%) was used to select cells with the highest expression level of a specific gene. A Ï2 test (scipy chi2_contingency) was performed per gene to identify if the selected cells were over-represented in a specific cell type (cell_type_level_4_explore), indicating the gene to be a marker gene. Genes that were predicted to be expressed only in a single cell type (Ï2 αâ=â1âÃâ10â50) were considered to be SGs and used for further analyses.
IBEX clinical cohort details and sample preparation
Human thymus samples were obtained from the pathology department of the Childrenâs National Medical Center in Washington, DC, after cardiothoracic surgery from children with congenital heart disease, as thymic tissue is routinely removed and discarded to gain adequate exposure of the retrosternal operative field. Use of these thymus samples for this study was determined to be exempt from review by the NIH Institutional Review Board in accordance with the guidelines issued by the Office of Human Research Protections. There were no genetic concerns for the patients in this cohort. Details about the cohort can be found in Supplementary Table 2. Human thymi were placed in PBS on receipt and processed within 24âh after surgery. Excess fat and connective tissue were trimmed and sectioned into <5âmm cubes. For IBEX imaging, human thymi were fixed with BD CytoFix/CytoPerm (BD Biosciences) diluted in PBS (1:4) for 2âdays. After fixation, all tissues were washed briefly (5âmin per wash) in PBS and incubated in 30% sucrose for 2âdays before embedding in OCT compound (Tissue-Tek) as described previously17,53.
IBEX sample imaging preparation
IBEX imaging was performed on fixed frozen sections as described previously17,53. In brief, 20âμm sections were cut on a CM1950 cryostat (Leica) and adhered to two-well chambered cover glasses (Lab-tek) coated with 15âμl of chrome alum gelatin (Newcomer Supply) per well. Frozen sections were permeabilized, blocked and stained in PBS containing 0.3% Triton X-100 (Sigma-Aldrich), 1% bovine serum albumin (Sigma-Aldrich) and 1% human Fc block (BD Biosciences). Immunolabelling was performed with the PELCO BioWave Pro 36500-230 microwave equipped with a PELCO SteadyTemp Pro 50062 Thermoelectric Recirculating Chiller (Ted Pella) using a 2-1-2-1-2-1-2-1-2 program. The IBEX thymus antibody panel can be found in Supplementary Table 3 and has been formatted as an Organ Mapping Antibody Panel54 (OMAP-17) accessible online (https://humanatlas.io/omap). Custom antibodies were purchased from BioLegend or conjugated in house using labelling kits for Lumican (AAT Bioquest, 1230) and LYVE-1 (Thermo Fisher Scientific, A20182). A biotin avidin kit (Abcam, ab64212) was used to block endogenous avidin, biotin and biotin-binding proteins before streptavidin application. Cell nuclei were visualized with Hoechst 33342 (Biotium) and the sections were mounted using Fluoromount G (Southern Biotech). Mounting medium was thoroughly removed by washing with PBS after image acquisition and before chemical bleaching of fluorophores. After each staining and imaging cycle, the samples were treated with two 15âmin treatments of 1âmgâmlâ1 of LiBH4 (STREM Chemicals) prepared in deionized H2O to bleach all fluorophores except Hoechst.
IBEX image acquisition and alignment
Representative sections from different tissues were acquired using the inverted Leica TCS SP8 X confocal microscope with a Ã40 objective (NA 1.3), 4 HyD and 1 PMT detectors, a white-light laser that produces a continuous spectral output between 470 and 670ânm as well as 405, 685 and 730ânm lasers. Panels consisted of antibodies conjugated to the following fluorophores and dyes: Hoechst, Alexa Fluor (AF)488, FITC, AF532, phycoerythrin (PE), eF570, AF555, iFluor 594, AF647, eF660, AF680 and AF700. All images were captured at an 8-bit depth, with a line average of 3 and 1024âÃâ1024 format with the following pixel dimensions: x (0.284âμm), y (0.284âμm) and z (1âμm). Images were tiled and merged using the LAS X Navigator software (LAS X v.3.5.7.23225). For IBEX tissue imaging, multiple tissue sections were examined before selecting a representative tissue section that contained several distinct lobules with multiple functional units, often resulting in an unusually shaped region of interest. Fluorophore emission was collected on separate detectors with sequential laser excitation of compatible fluorophores (3â4 per sequential) used to minimize spectral spillover. The Channel Dye Separation module within LAS X v.3.5.7.23225 (Leica Microsystems) was then used to correct for any residual spillover. For publication-quality images, Gaussian filters, brightness/contrast adjustments and channel masks were applied uniformly to all images. Image alignment of all IBEX panels was performed as described previously using SimpleITK55,56. Additional details on antibodies, protocols and software can be found on the IBEX Knowledge-Base (https://doi.org/10.5281/zenodo.7693279).
IBEX 3D single nuclei segmentation with cellpose
IBEX images were converted from .ims format (Imaris, Oxford Instruments, v.9.5.0 and v.10.0.0) to 3D stacks (TIFF) by individual channels with FIJI v.1.54j. We then applied a custom pipeline for 3D single nuclei segmentation with cellpose v.2.1.1:
-
(1)
Image preparation: individual TIFF images were separated by z plane and channel and the sample channel metadata were extracted as a .csv file. The nuclear staining channel (Hoechst) was located, and a set of tiled 3D image arrays was generated.
-
(2)
Segmentation: the image arrays were sequentially segmented with cellpose using specific parameters, for example, diameter, resampling, anisotropy, thresholds and batch size. We used tiles to overcome restrictions on GPU and RAM resources.
-
(3)
Restitching and formatting: after segmentation, tiles were stitched back together, and all segmentation masks were stored in a uint32-bit image format to hold more than 65,535 mask labels, as a typical sample produces 200,000â700,000 cell masks.
-
(4)
Handling high nuclear density: the high nuclear density, especially in the thymic cortex, produced scenarios in which segmentation masks were often bordered by neighbouring cells, resulting in substantial inter-cell signal bleed. To overcome this issue, we excluded pixels in the interface between two cells such that the mask boundary was preserved if no neighbouring cell was present.
-
(5)
Mask filtering: we removed small cell masks to avoid noise and cell fragments using skimage.morphology.remove_small_objects.
-
(6)
Signal intensity extraction: for each cell mask post-filtering, we extracted the mean and maximum signal intensity for each channel in the IBEX image. This produced a cellXprotein file for each sample.
-
(7)
Data merging: once all sample single-cell segmentations were collected, we merged all samples (nâ=â8) and stored metadata, channel and spatial information in a unified AnnData object, totalling more than 1.1âmillion cells.
Label transfer to IBEX cells from scRNA-seq and CITE-seq reference atlas with ISS patcher
We used a KNN algorithm to compare the annotated cell types in our scRNA-seq reference atlas with the IBEX single-cell segmentations. For this purpose, the protein expression in the IBEX query cells was matched with the RNA expression of the corresponding genes in the scRNA-seq reference. Protein and gene names were matched according to Supplementary Table 11. Batch effects were removed from IBEX with scanpy.pp.combat(ibex_gene, key=âsampleâ, inplace=True). We next subsetted each IBEX sample and ran the KNN prediction algorithm per sample with kâ=â30, including the following steps:
-
(1)
The shared feature space between the two objects was identified, log-normalized and z-scored on a per-object basis.
-
(2)
The KNN of the low-dimensional observations (IBEX) in the high-dimensional space (GEX) were identified. The counts of absent features were imputed as the mean of the high-dimensional neighbours.
-
(3)
On the basis of majority voting, the most frequent cell annotation in the GEX reference was assigned to the IBEX query cell. The proportion of KNNs that contributed to the majority voting was recorded as the evidence fraction (KNNf). For example, if out of the 30 nearest neighbours, 13 were labelled as A, 10 as B and 7 as C in GEX, then the IBEX cell received the label A. The fraction in this case refers to the proportion of the 30 nearest neighbours that contributed to the majority label, which would be KNNf = 13/30â=â0.43 for label A.
In some of our IBEX samples AIRE staining in particular produced a relatively high level of non-specific signal and low signal-to-noise ratio. As a consequence, we identify some predicted AIRE+ mTECII cells that are not in the medulla and have capsular/cortical localization (Fig. 4g). We had cases of individual samples for which a specific antibody did not perform well or was missing (for example, for IBEX_01 the KRT10 antibody was out of stock). However, as ISS patcher was run on each sample separately, this did not affect proper scaling of that marker and its use of it for mapping in other samples. Moreover, by selecting cells with a higher proportion of matched KNN cells from the single-cell data (KKNf) these effects are reconciled through the removal of cells with low-confidence mapping. We would like to flag this point for future researchers who want to reuse our datasets.
The general code for the ISS patcher KNN mapping algorithm can be found in the dedicated GitHub repository (https://github.com/Teichlab/iss_patcher/tree/main) and the full example for KNN mapping using IBEX is reported in the GitHub repository for this Article (see Code availability).
To annotate T cell types in the IBEX data with high accuracy, we applied the ISS patcher with the CITE-seq T lineage data as a reference as follows: we first applied the KNN algorithm to IBEX data using the scRNA-seq reference atlas to identify and subset IBEX T lineage cells. We then used the CITE-seq data as a reference to repeat the KNN-based annotation on these selected cells. This KNN-based reannotation was performed on a hybrid RNA/protein reference, which included protein measurements for the 19 markers assayed in both CITE-seq and IBEX in addition to RNA measurements for the remaining 23 genes as described in Supplementary Table 11. We used the same KNN implementation as described above but with kâ=â7, while also imputing CD4 and CD8 pseudotime.
Human thymus FFPE processing
Human paediatric samples were obtained from cardiac corrective surgeries. Removed thymi were directly moved to HypoThermosol (Sigma-Aldrich, H4416-100ML), shipped with a courier with ice packs and processed in under 24âh after surgery. After arrival, for FFPE processing, the samples were cut to approximately 1âcm2 pieces with sharp scissors in 1à DPBS. Tissue pieces were then rinsed in clean DPBS to remove any excess HypoThermosol, patted dry with wipes (Kimtech) and placed into 10% formalin (cellpath, BAF-6000-08A) for 16â24âh at room temperature. The next day, tissues were dehydrated and embedded in wax, then kept at 4â°C.
RareCyte immunostaining and 14-plex imaging
Multiplex immunofluorescence and single round imaging was performed as described previously57. All steps were performed at room temperature unless stated otherwise. In brief, FFPE blocks were sectioned using a microtome (Leica, RM2235) at 3.5â5âµm thickness and placed onto a superfrost slide (Thermo Fisher Scientific, 12312148). Slides were dried at 60â°C for 60âmin to ensure that tissue sections had adhered to the slides. Tissue sections were deparaffinized and subjected to antigen retrieval using the BioGenex EZ-Retriever system (95â°C for 5âmin followed by 107â°C 5âmin). For OCT sections, 7âµm sections were taken using a cryostat (Leica CM3050S), placed onto SuperFrost Plus slides (VWR) and immediately submerged in 10% buffered saline formalin (Cellpath, BAF-6000-08A) for 1âh at room temperature. The samples were then subjected to the following steps similarly to the FFPE samples. To remove autofluorescence, slides were bleached with AF quench buffer (4.5% H2O2, 24âmM NaOH in PBS). The slides were quenched for 60âmin using the âhighâ setting with a strong white-light exposure followed by further quenching for 30âmin using the 365ânm âhighâ setting using a UV transilluminator. The slides were rinsed with 1à PBS and incubated in 300âµl of Image-iT FX Signal Enhancer (Thermo Fisher Scientific, I36933) for 15âmin. The slides were rinsed again and 300âµl of labelled primary antibody staining cocktail was added to the tissue, which was subsequently incubated for 120âmin in the dark within a humidity tray. All antibodies were prediluted according to company recommendations and were not adjusted further (Supplementary Table 4). The slides were washed with a surfactant wash buffer and 300âµl of nuclear staining in goat diluent was added to the slide. The slides were then incubated in the dark for 30âmin in a humidity tray. The slides were then washed and placed in 1à PBS. Finally the slides were coverslipped using ArgoFluor mount medium and left in the dark at room temperature overnight to dry. The slides were imaged the next day using the RareCyte Orion microscope with a Ã20 objective and relevant acquisition settings were applied using the software Artemis v.4.
RNAscope processing and imaging
For RNAscope analysis, thymus tissue was processed as described above for Visium sectioning. Sections were cut from the fresh frozen OCT-embedded (OCT, Leica) samples at a thickness of 10âμm using a cryostat (Leica, CM3050S) and placed onto SuperFrost Plus slides (VWR). Sections were stored at â80â°C until staining. The sections were removed from the â80â°C storage and submerged in chilled (4â°C) 4% PFA for 15âmin, then acclimatized to room temperature 4% PFA over 120âmin. The sections were then briefly washed in 1à PBS to remove any remaining OCT. Then, the sections were dehydrated in a series of 50%, 70%, 100% and 100% ethanol (5âmin each) and air-dried before performing automated 4-plex RNAscope.
Using the automated Leica BOND RX, RNAscope staining was performed on the fresh frozen sections using the RNAscope LS multiplex fluorescent Reagent Kit v2 Assay and RNAscope LS 4-Plex Ancillary Kit for LS Multiplex Fluorescent (Advanced Cell Diagnostics (ACD), Bio-Techne) according to the manufacturerâs instructions. All of the sections were subjected to 15âmin of protease III treatment before staining protocols were performed. Before running RNAscope probe panels, the RNA quality of fresh frozen samples was assessed using multiplex positive (RNAscope LS 2.5 4-plex Positive Control Probe, ACD Bio-Techne, 321808) and negative (RNAscope 4-plex LS Multiplex Negative Control Probe, ACD Bio-Techne, 321838) controls.
The probes were labelled using Opal 520, 570 and 650 fluorophores (Akoya Biosciences, 1:1,000) and one probe channel was labelled using Atto 425-streptavidin fluorophore (Sigma-Aldrich, 1:500), which was first incubated with TSAâbiotin (Akoya Biosciences, 1:400). The following RNAscope 2.5 LS probes were used for this study: Hs-AIRE (ACD Bio-Techne, 551248), Hs-LY75-C2 (ACD Bio-Techne, 481438-C2), Hs-CAMP-C3 (ACD Bio-Techne, 446248-C3), Hs-EPCAM-C4 (ACD Bio-Techne, 310288-C4), Hs-IGFBP6-C1 (ACD Bio-Techne, 496068) and Hs-DLK2-C3 (ACD Bio-Techne, 425088-C3). All nuclei were DAPI stained (Life Technologies, D1306). Details are provided in Supplementary Table 12.
Confocal imaging was performed on the Perkin Elmer Operetta CLS High Content Analysis System using a Ã20 (NA 0.16, 0.299âμmâpxâ1) water-immersion objective with 9-11 z-stacks with 2âµm step. Channels: DAPI (excitation, 355â385ânm; emission, 430â500ânm), Atto 425 (excitation, 435â460ânm; emission, 470â515ânm), Opal 520 (excitation, 460â490ânm; emission, 500â550ânm), Opal 570 (excitation, 530â560ânm; emission, 570â620ânm), Opal 650 (excitation, 615â645ânm; emission, 655â760ânm). Confocal image stacks were stitched as individual z stacks using proprietary Acapella scripts provided by Perkin Elmer, and visualized using OMERO Plus (Glencoe Software).
The contrast used for Extended Data Fig. 6e was as follows: DLK2 (magenta 150â500), IGFBP6 (yellow 200â1500), LY75 (green 200â4000), EPCAM (red 300â2500).
CITE-seq tissue processing
Paediatric thymus samples from children undergoing cardiac surgery were obtained according to and used with the approval of the Medical Ethical Commission of Ghent University Hospital, Belgium (EC/2019-0826) through the haematopoietic cell biobank (EC-Bio/1-2018). Thymus tissue was cut into small pieces using scalpels and digested with 1.6âmgâmlâ1 collagenase (Gibco, 17104-019) in IMDM medium for 30âmin at 37â°C with regular agitation to generate a single-cell suspension. The reaction was quenched with 10% FBS and the thymocyte suspension was passed through a 70âμm strainer to remove undigested tissue. Cells were frozen in FBS containing 10% DMSO and stored in liquid nitrogen until needed.
CITE-seq antibody preparation
The TotalSeq-C Human Universal Cocktail 1.0 (BioLegend, 399905) was resuspended according to the manufacturerâs instructions. In brief, the lyophilized cocktail was equilibrated to room temperature for 5âmin and then centrifuged at 10,000g for 30âs. Then, 27.5âµl cell staining buffer (BioLegend, 420201) was added and the tube was vortexed for 10âs, incubated for 5âmin at room temperature and vortexed again. The resuspended cocktail was centrifuged for 30âs at room temperature at 10,000g and the entire volume was transferred to a low-bind tube. Finally, the tube was centrifuged again at 14,000g for 10âmin at 4â°C and 25âµl of the supernatant was used per sample (2âÃâ106 cells in 200âµl).
In total, 13 TotalSeq-C antibodies (BioLegend) were titrated individually by flow cytometry using PE-conjugated versions of the same antibody clone as recommended by BioLegend. After choosing a suitable concentration for each antibody, a master mix was prepared for cell staining. For this, all antibodies were initially diluted in the cell staining buffer to obtain a concentration 100-fold higher than the desired final staining concentration. Then, 2âµl of each diluted antibody substock was combined in a master mix, which was added to the cells for labelling in a total volume of 200âµl. Details on the TotalSeq-C antibodies are provided in Supplementary Table 6.
CITE-seq sample preparation
Cells were thawed slowly by gradually adding 15 volumes of pre-warmed IMDM medium and pelleted at 1,700ârpm for 6âmin at 4â°C. After resuspending in PBS, cells were passed through a 70âμm strainer to remove clumps. Enrichment for viable cells was achieved using a magnetic bead-based dead cell removal kit (Miltenyi, 130-090-101). For this, cells were pelleted as before, washed with 1à binding buffer (part of the kit, prepared with sterile distilled water) and resuspended in dead cell removal microbeads (part of the kit) at a concentration of 107 cells per 100âµl beads. After incubation at room temperature for 15âmin, cells were applied to an LS column (Miltenyi, 130-122-729), which was prerinsed with 3âml 1à binding buffer. The column was washed four times with 3âml binding buffer and the flow-through containing viable cells was collected. Cells in the flow-through were pelleted and viability was confirmed using trypan blue. A total of 2âÃâ106 viable cells was used for TotalSeq-C and anti-CD3-PE antibody staining. For this purpose, cells were washed with cell staining buffer (BioLegend, 420201), pelleted at 600g for 10âmin at 4â°C and resuspended in 90âµl cell staining buffer. Then, 10âµl Human TruStain FcX blocking solution (BioLegend, 422301) was added and cells were incubated for 10âmin at 4â°C. The TotalSeq-C Human Universal Cocktail 1.0 (BioLegend, 399905) was resuspended as described above, centrifuged at 14,000g for 10âmin at 4â°C and 25âµl of the supernatant was added to the blocked cells. Individual TotalSeq-C antibodies were prepared as described above and 26âµl of the master mix was added to each sample. To facilitate enrichment of immature and mature thymocytes by FACS, 10âµl anti-CD3-PE (SK7, BioLegend, 344805) was added and the samples were topped up with 40âµl cell staining buffer resulting in a total staining volume of 200âµl. The samples were incubated for 30âmin at 4â°C in the dark. To wash off unbound antibodies, cell staining buffer was added to the samples, and cells were pelleted for 10âmin at 600g at 4â°C. All supernatant was removed, cells were resuspended in cell staining buffer, transferred to a new tube and pelleted as before. Cells were again resuspended in cell staining buffer and pelleted and this wash step was repeated once more before cells were resuspended in 200âµl MACS buffer (2% FCS, 2âmM EDTA in PBS) in preparation for sorting. Then, 1âµl propidium iodide (Invitrogen, 230111) was added for detection of dead cells and samples were sorted on the BD FACSAria III or BD FACSAria Fusion cell sorter using a 100âμm nozzle and a maximum flow rate of 4 (FACSDiva v.8.0.2, reanalysis with FlowJo v.10.8.2). Cells were gated using forward/side scatter to remove doublets and debris, then dead cells were excluded based on PI staining. CD3â and CD3+ cells were collected separately in cooled IMDMâ+â50% FCS (Supplementary Fig. 19b). After completion of the sort, collection tubes were topped up with DPBS and cells were pelleted at 400g for 10âmin at 4â°C. The supernatant was removed and cells were resuspended at an estimated concentration of 1,500 cells per µl in PBSâ+â0.04% BSA (Miltenyi, 130-091-376), of which 16.5âµl was used in the GEM generation step. The Next GEM Single Cell 5â² Kit v2 (10x Genomics, 1000265) was used to prepare the reaction master mix, and load cells, gel beads and partitioning oil on a Chip K (10x Genomics, 1000286) according to the manufacturerâs protocol CG000330 Rev A. GEMs were generated using a Chromium Controller (10x Genomics), transferred to a tube strip and reverse transcription was performed in a BioRad C1000 Touch Thermal Cycler according to the protocol. The samples were stored at 4â°C overnight and the library preparation was carried out the next day.
CITE-seq library preparation and sequencing
Feature barcode (FB), gene expression (GEX) and TCR libraries were prepared according to protocol CG000330 Rev A (10x Genomics) using the Chromium Next GEM Single Cell 5â²GEM Kit v2 (10x Genomics, 1000244), Library Construction Kit (10x Genomics, 1000190), 5â² Feature Barcode Kit (10x Genomics, 1000256), Human TCR Amplification Kit (10x Genomics, 1000252), Dual Index Kit TT set A (10x Genomics, 1000215) and Dual Index Kit TN set A (10x Genomics, 1000250). The protocol version for >6,000 cells was followed and libraries were amplified for 13 cycles (cDNA), 14 cycles (GEX), 8 cycles (FB) or 12â+â10 cycles (TCR libraries). Library quality and quantity were checked on the Bioanalyzer instrument (Agilent) using a high-sensitivity DNA assay. Libraries were pooled and sequenced on the NovaSeq 6000 instrument (Illumina) to a minimum of 25,000 reads per cell for GEX, 10,000 reads per cell for FB and 5,000 reads per cell for TCR libraries.
CITE-seq quality control and denoising
CITE-seq data were processed using the R packages Seurat58 (v.4.3.0), SeuratObject (v.4.1.4), SeuratDisk (v.0.0.0.9021), SingleCellExperiment (v.1.24.0), Matrix (v.1.6-4), matrixStats (v.1.2.0), dplyr (v.1.1.4), tidyr (v.1.3.1), reshape2 (v.1.4.4), BiocNeighbors (v.1.20.2), BiocParallel (v.1.36.0), stringr (V.1.5.1), reticulate (v.1.35.0) and sceasy (v.0.0.7). Data were visualized using ggplot2 (v.3.5.0), ggrastr (v.1.0.2), ggridges (v.0.5.6) and RColorBrewer (v.1.1-3).
Fastq files from FB libraries were mapped with Cell Ranger v.7.0.0. GEX libraries were mapped with STARsolo as described above for scRNA-seq data. CITE-seq data were first subjected to quality-control processing based on RNA properties as described above. For the retained high-quality cells, ADT data were then denoised using dsb (v.1.0.3)59. For this purpose, empty droplets were identified in the unfiltered mapping output and used as a reference for estimating noise and antibody background levels. Approximately 1.4âmillion droplets were selected on the basis of RNA countâ<â240 reads, ADT count between 120 and 350 reads, and <5% mitochondrial reads to ensure that damaged cells were not included in the subset. During denoising and normalization with DSBNormalizeProtein, ADT data from droplets were used for background correction and the seven isotype control antibodies included in the TotalSeq-C Human Universal Cocktail were used to determine technical variation. Data were scaled based on the background by subtracting the mean and then dividing by the standard deviation of the empty droplets. Negative values after denoising, which correspond to very low expression, were set to zero to improve interpretation and visualization. In an additional quality-control step, cells in which less than 100 of the antibodies were detected were removed from the dataset as technical artefact. Moreover, a subset of cells affected by aggregates of the antibodies against TCRγδ, CD199 (CCR9), CD370, CD357 and XCR1 was removed from the dataset due to unreliable surface-staining properties.
CITE-seq annotation
Annotation of the CITE-seq data was carried out on integrated RNA and denoised ADT modalities. For this purpose, both data modalities were log-normalized and the top 2,000 highly variable genes were identified, followed by PCA on the scaled highly variable genes using the standard functions in the Seurat package. MNN correction was applied to the PCA loadings matrix using the reducedMNN function from the Batchelor package60 (v.1.18.1) to reduce batch and donor effects between samples. To integrate both modalities, multimodal neighbours and modality weights were identified and a weighted nearest neighbours (WNN) graph58 was constructed using FindMultimodalNeighbors based on the PCA. The number of PCs to be used was determined based on the difference in variation of consecutive PCs being higher than 0.1%. A UMAP visualization was generated based on the WNN graph to represent the weighted combination of both modalities. Furthermore, a supervised PCA (sPCA) was performed on transcriptome data using the RunSPCA function of the Seurat package to obtain dimensionality reduction for the RNA modality that represents the structure of the WNN graph58.
To identify cell types and developmental stages, we performed Leiden clustering at a low resolution based on the sPCA using the Seurat functions FindNeighbors and FindClusters. The obtained clusters were then subsetted and analysed individually starting with the most immature cluster, which was identified based on high expression of the surface marker CD34. For each subset, normalization and scaling for RNA and ADT data were repeated as described above and a new WNN UMAP and sPCA were constructed. A combination of Leiden clustering on the sPCA and thresholding of protein levels was used to identify cell types. Moreover, to identify proliferating cells, cell cycle scoring was performed using the G2M and S phase markers supplied in the Seurat package. The FindMarkers function (Seurat) and the package singleCellHaystack61 (v.1.0.0) were used to identify differentially expressed genes and surface markers in a cluster-based and cluster-independent manner, respectively. Distinction of CD4 and CD8 lineage maturation stages was based on CD45RA, CD45RO and CD1a and identical expression cut-offs were used for both lineages to ensure that subsets would be directly comparable.
Paired TCR-seq data were processed with Dandelion47 v.0.3.1 and information about productive or non-productive TRA and TRB rearrangements was extracted for each cell to validate cell type annotations after clustering.
CITE-seq pseudotime analysis
To carry out trajectory inference for αβT lineage cells, CITE-seq data were subsetted to contain DP_pos_sel, DP_4hi8lo, SP_CD4_immature, SP_CD4_semi-mature, SP_CD4_mature, SP_CD8_immature, SP_CD8_semi-mature and SP_CD8_mature cells. A new WNN UMAP was constructed based on surface protein and RNA. Slingshot62 (v.2.6.0) was used to establish a minimum spanning tree on the WNN UMAP using the getLineages function based on mutual nearest neighbour-based distance with DP_pos_sel set as start point and SP_CD4_mature and SP_CD8_mature specified as end points. Smooth lineages were obtained using the getCurves function and the derived pseudotime orderings were used to assess transcript and surface marker expression throughout differentiation.
CITE-seq cell2location mapping, integration and processing
For the focused multimodal analysis, we subsetted the full scRNA-seq dataset to include only paediatric data and further removed T lineage cells without CITE-seq protein information. We then performed cell2location as described above to obtain deconvolved cell type mapping based on CITE-seq annotations. In addition, as cell2location deconvolves spots by unique pseudobulk expression profiles from a single-cell reference based on discrete cell annotations, the mapping resolution is limited to that of the annotated cell subsets. To investigate the continuous spatiotemporal nature of CD4/CD8 cell lineages at increased resolution, we performed high-resolution Leiden clustering on the CITE-seq data using scanpy (resolution=35), resulting in 567 cell clusters. These cell clusters were then mapped to our paediatric Visium data with cell2location as described above. To measure the position of each cell cluster across the CMA, we selected Visium spots with the highest cluster label abundance (above percentile 95%) and calculated the weighted mean CMA values for these spots. This value was then assigned this to the cells comprising the associated cluster in the single-cell object. Further details are provided in the GitHub repository for this Article (see Code availability).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.