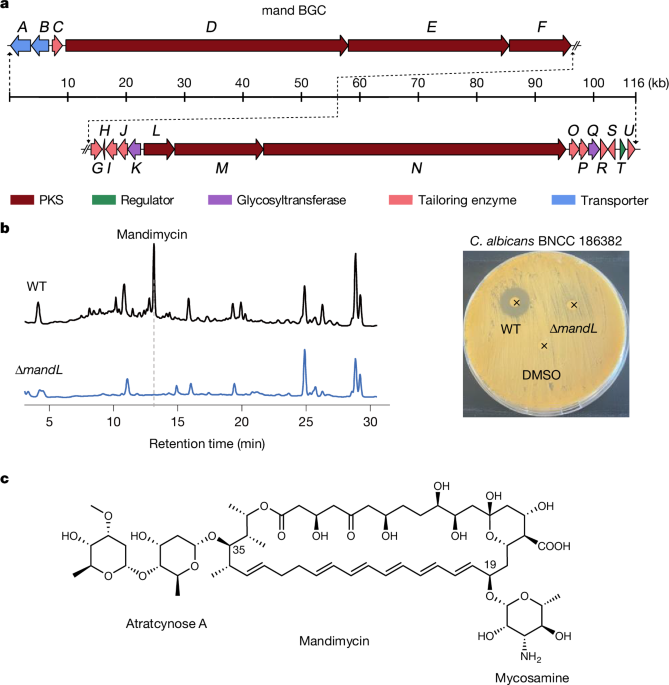
Identification and bioinformatic analysis of mandimycin BGC
To identify potential BGCs encoding previously unknown mycosamine-containing polyene macrolide antibiotics, a localized MiSM database was constructed using microbial genome sequencing data, including ‘complete’, ‘chromosome’, ‘scaffold’ and ‘contig’ assemble levels, from The National Center for Biotechnology Information. Moreover, 150 isolated Streptomyces genomes sequenced in our laboratory were incorporated into the MiSM database. Next, putative BGCs in these genomes were predicted and annotated using antiSMASH v.7.0 with four extra features (that is, known cluster blast, subcluster blast, MiBiG cluster comparison and active site finder)11. The identified BGCs were organized in a local server using the MySQL model under the DB-API database frame. A customized profile Hidden Markov Model (pHMMer)43 was built using 11 known glycosyltransferase sequences detailed in Supplementary Table 1 and used to scan against the localized MiSM database. A cut-off e-value of 5e−181 was used to identify sequences encoding glycosyltransferases responsible for transferring the mycosamine motif onto the polyene macrolide ring of potential polyene macrolide antibiotics. Candidate sequences were subsequently trimmed and aligned using the Molecular Evolutionary Genetics Analysis v.11 (MEGA11) software with the Muscle algorithm44. A phylogenetic tree was constructed using the neighbour-joining method and visualized on the interactive Tree Of Life (iTOL) website45.
Knockout of mandL and mandQ in S. netropsis
To validate the mand BGC and obtain the bis-deglycosylated counterpart, we constructed targeted deletion mutants of the polyketide synthase gene (ΔmandL) and the di-digitoxose glycosyltransferase gene (ΔmandQ). In brief, two fragments, one upstream and one downstream of the targeted gene, were PCR-amplified from the genomic DNA of S. netropsis DSM 40259 using primer pairs MandL_KOUF/MandL_KOUR and MandL_KODF/MandL_KODR for mandL and MandQ_KOUF/MandQ_KOUR and MandQ_KODF/MandQ_KODR for mandQ, respectively. These amplicons were then cloned into the vector pKC1139 digested with BamHI for mandL or XbaI for mandQ, yielding the plasmids pKC1139-MandL_KO and pKC1139-MandQ_KO. These constructs were subsequently introduced into S. netropsis DSM 40259 through Escherichia coli–Streptomyces conjugation, and apramycin-resistant exconjugants were selected and incubated in mannitol soya flour (MS) medium at 30 °C to generate double-crossover mutants. Deletion of mandL and mandQ was verified by PCR analysis using the primer pairs MandL_KOUF/MandL_KODR and MandQ_KOTF/MandQ_KOTR, respectively. The resulting PCR products were further confirmed by Sanger sequencing.
Fermentation of S. netropsis and purification of mandimycin
Spores of S. netropsis DSM 40259 were generated by culturing on ISP4 agar plates at 30 °C for 7 days. Spores were aseptically inoculated into a 50 ml starting culture of tryptic soy broth in 250-ml baffled Erlenmeyer flasks using sterile inoculum sticks. The flasks were incubated with agitation at 30 °C for 2 days. Next, 0.5 ml of the tryptic soy broth seed culture was used to inoculate 50 ml of F2 medium (glucose 69 g l−1, beef extract 25 g l−1, CaCO3 9 g l−1 and KH2PO4 0.1 g l−1) in 250 ml baffled Erlenmeyer flasks. Mandimycin B was fermented in FS/9 medium (soybean meal 30 g l−1, glucose 40 g l−1 and CaCO3 10 g l−1). The flasks were then agitated at 30 °C, 200 rpm for 10 days. The cultures, including the mycelia, were extracted overnight using n-butanol at a 1:1 ratio. The resulting organic extract was separated and concentrated using a rotary evaporator. The concentrated extracts were then fractionated on a YMC-GEL C18 column using a stepwise gradient elution of methanol/water (10%, 30%, 50%, 70%, 90% and 100% methanol). The fractions containing mandimycin were identified using ultraperformance liquid chromatography–mass spectrometry (Waters), and the pure form of mandimycin was further purified through reverse-phase high-performance liquid chromatography (HPLC; Shimadzu, ShimNet HE C18-AQ, 5 μm optimal bed density (OBD), 19 × 250 mm, 3 ml min−1 from 30% to 90% acetonitrile in water over 60 min). The purity and identity of mandimycin were confirmed by ultraperformance liquid chromatography–mass spectrometry. The target HPLC fractions were combined and lyophilized, yielding a pure powder for subsequent biological assays.
Structure elucidation of mandimycin and mandimycin B
To elucidate the structures of mandimycin and mandimycin B, the purified compounds were analysed by NMR spectroscopy (Bruker AVANCE NEO 600 MHz and Bruker AVANCE III HD 700 MHz) and HR-ESI–MS (Waters-Gs-XS-QTOF and Thermo Scientific Q Exactive Orbitrap). Mandimycin or mandimycin B (around 20 mg for each) was dissolved in dimethyl sulfoxide (DMSO)-d6 for one-dimensional (1H and 13C) and two-dimensional (HSQC, HMBC, 1H-1H COSY, TOCSY, NOESY) NMR analysis. (HSQC, heteronuclear single quantum coherence; HMBC, heteronuclear multiple bond correlation; COSY, correlation spectroscopy; TOCSY, total correlation spectroscopy; NOESY, nuclear Overhauser effect spectroscopy). NMR data were acquired on either a Bruker AVIII 600 MHz instrument equipped with a cryoprobe or a Bruker AVIII 700 MHz instrument. Chemical shifts are reported in parts per million (ppm) relative to tetramethyl silane using the residual solvent signals at 2.50 ppm in 1H NMR and 39.5 ppm in 13C NMR as internal signals. HR-ESI-MS data were acquired on an Exactive Orbitrap mass spectrometer under both positive and negative ionization modes by direct infusion. The acquired data were further processed using Xcalibur software. A detailed interpretation of NMR and HR-ESI–MS data for the structure elucidation of mandimycin and mandimycin B is provided in the Supplementary Information.
MIC assay
The in vitro antimicrobial potency of mandimycin and mandimycin B was assessed by determining their MIC values following the guidelines of the Clinical and Laboratory Standards Institute19. Mandimycin was tested against a panel of fungal pathogens as well as Gram-positive and Gram-negative bacterial pathogens, as detailed in Supplementary Table 4. The assays were performed in 96-well microlitre plates, with amphotericin B, vancomycin and ciprofloxacin serving as positive controls for fungal, Gram-positive and Gram-negative pathogens, respectively. All test compounds were dissolved in sterile DMSO (SCR, CN) to achieve a concentration of 12.8 mg ml−1. Assay strains were cultured from a single colony overnight and subsequently diluted 5,000-fold in fresh Luria Bertani (LB) (bacteria) or yeast peptone dextrose (YPD) (fungi) medium. The microbial suspension was subsequently distributed into the first row (100 µl per well) and the rest of the rows (50 µl per well). The test compounds were serially diluted across 96-well plates using a twofold serial dilution in a volume of 50 µl to give a range of final concentrations of each compound between 64 and 0.125 μg ml−1. To avoid edge effects, the top and bottom rows of each plate were filled with 100 µl of medium. Plates were incubated at 37 °C (bacteria) or 30 °C (fungi) for 16 h. The lowest concentration at which there was no visible growth of bacteria or fungi was recorded as the MIC. All assays were performed in duplicate and repeated three independent times.
Cytotoxicity assay
The cytotoxicity of mandimycin and mandimycin B was evaluated using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay46. HepG2 (human hepatoblastoma cell line), HK-2 (human renal proximal tubule epithelial cell line), SK-Hep-1 (human hepatic adenocarcinoma cell line) and PANC-1 (human pancreatic cancer cell line) cells were cultured in DMEM supplemented with 10% fetal bovine serum until the exponential phase was reached. RPTEC cells were cultured in renal epithelial cell basal medium supplemented with a renal epithelial cell growth kit until the exponential phase was reached. Subsequently, cells were seeded into a 96-well, flat-bottom microlitre plate at a density of 2,500 cells per well. After 24-h incubation at 37 °C with 5% CO2, the medium was aspirated and replaced with 100 μl of fresh medium containing mandimycin or mandimycin B at concentrations ranging from 128 μg ml−1 to 0.25 μg ml−1. After an additional 48-h incubation, the culture medium was removed, and 110 μl of MTT solution (10 μl of 5 mg ml−1 MTT in PBS premixed with 100 μl of DMEM) was added into each well. The mixture was incubated for an additional 3 h at 37 °C to enable formazan crystal formation, followed by solubilization with 100 μl of solubilization solution (40% dimethylformamide, 16% sodium dodecyl sulfate and 2% acetic acid in H2O). The absorbance of each well was measured at an optical density at 570 nm using a microplate reader (Epoch Microplate Spectrophotometer, BioTek). The half-maximal inhibitory concentration values of mandimycin and mandimycin B were calculated (GraphPad Prism 9) as the concentration of compound required to inhibit 50% of cell growth, relative to control wells without any compound. All experiments were performed in three biological replicates.
Feeding assay
The impact of fungal cell membrane components on the antifungal activity of mandimycin was evaluated using C. albicans BNCC 186382. Membrane components including ergosterol, cholesterol, lanosterol, phosphatidylcholine, phosphatidylethanolamine, phosphatidylserine, phosphatidylinositol, phosphatidylglycerol, cardiolipin, sphingomyelin, phosphorylcholine, phosphoserine, phosphorylethanolamine, β-1,3-glucan, β-1,6-glucan and mannose were prepared in 10% sterile DMSO. Each solution (10 μl) was added to individual wells of 96-well microtitre plates, resulting in final concentrations of these cell membrane components of 0.05, 0.1, 0.2, 0.4, 0.8 and 1.0 mg ml−1. The MIC value of each treatment against C. albicans was recorded using the same method described for the MIC assay. The experiment was conducted in three biological replicates.
ITC assay
The binding of mandimycin to phospholipids (phosphatidylcholine, phosphatidylinositol, phosphatidylserine, cardiolipin, sphingomyelin and phosphatidylglycerol) and sterols (ergosterol and cholesterol) was measured using ITC. LUVs with a size of 0.1 μm were prepared by mixing DOPC (Avanti Polar Lipids) and 5% of each phospholipid or sterol in chloroform (1:1). The resulting lipid suspension was evaporated to dryness in vacuo to yield a lipid film, followed by rehydration using 5 mM HEPES (pH 7.4, 100 mM NaCl). Subsequently, the hydrated sample was extruded ten times through a 0.1-μm filter membrane using an Avanti Mini Extruder. ITC experiments were performed on a PEAQ-ITC instrument at 25 °C using a solution of 1 mM of mandimycin, mandimycin B or amphotericin B along with 600 µM DOPC LUVs. The titration process involved an initial injection of 0.23 μl, followed by 18 injections of 2 μl at 80-s intervals, with continuous stirring at 500 rpm. Data were analysed using PEAQ-ITC software, and the thermodynamic parameters (enthalpy (ΔH), entropy (ΔS) and the equilibrium binding constant (apparent Kd)) were calculated using a one-binding-site model.
UV–vis spectroscopy for sterol and phospholipid binding
To investigate the potential formation of complexes between mandimycin and mandimycin B with sterols or phospholipids, the UV–vis binding assay was carried out following established protocols27. A stock solution (100 mM) of each phospholipid (that is, phosphatidylinositol, phosphatidylcholine, phosphatidylethanolamine, phosphatidylglycerol, cardiolipin and sphingomyelin) and sterol (that is, ergosterol and cholesterol) was prepared in chloroform and subsequently diluted to a final concentration of 1 mM using DMSO. Mandimycin or mandimycin B was dissolved in DMSO to prepare a stock solution of 10 mM, which was further diluted to a working concentration of 1 mM using DMSO. Amphotericin B and DMSO were used as positive and negative controls, respectively. Complexes were prepared by mixing the stock solutions in various ratios to achieve a final volume of 1 ml. The mixture was then incubated at room temperature for 30 min. The UV–vis absorbance spectra ranging from 310 to 400 nm were recorded on a Thermo Scientific Multiskan SkyHigh instrument and plotted using Origin 2021 (v.9.8).
Isolating resistant mutants
Single colonies of various pathogenic fungal species (C. auris BNCC 357785, C. albicans BNCC 186382, C. neoformans BNCC 225501, C. glabrata BNCC 337348 and C. tropicalis BNCC 340288) were cultured overnight in 5 ml YPD broth at 30 °C with continuous shaking at 200 rpm. The overnight culture was diluted to a concentration of 109 cells per 100 µl using fresh YPD medium and plated onto an YPD agar plate containing 8× MIC of tested compounds (that is, mandimycin, amphotericin B, nystatin and natamycin). Plates were statically incubated at 30 °C for 2 days, and resistant colonies were observed and recorded. MIC values of selected resistant mutants were determined using the same method as described for the MIC assay.
In vivo nephrotoxicity assay
Specific-pathogen-free female Insitute of Cancer Research (ICR) mice, aged 6 weeks and weighing 23–27 g, were used in this study (Hangzhou Medical College, China). The mice were randomly housed in individual cages and divided into 12 groups with 4 mice per group. Mice were acclimatized for 3 days before the experiments. Amphotericin B or a solvent without any antibiotics was used as the positive control and placebo, respectively. Compounds were formulated in a solution containing 10% DMSO and 10% Tween 80. Subsequently, each group of mice received daily subcutaneous or intravenous injection of various concentrations of the tested compounds or a placebo. The protein concentrations of toxicity-related biomarkers, including KIM-1, TIMP-1, LCN-2 and SPP-1, were measured using commercial kits (Cloud-Clone), following the protocols provided. Gene expression levels of Kim1, Timp1, Lcn2 and Spp1 were assessed using reverse-transcription PCR with specific primer sets (Kim1F/R, Lcn2F/R, Timp1F/R and Spp1F/R), with Gadph as a control. The quantifications were performed using Applied Biosystems QuantStudio 3. Finally, all animals were euthanized, and the kidney tissues were collected, fixed, dissected and stained with haematoxylin and eosin (H&E). Pathological changes in tubular degeneration, necrosis, cellular casts, dilation, congestion and protein casts were evaluated and scored in a double-blinded fashion by a clinical pathologist. All animal study procedures were approved by Animal Ethics Committee of China Pharmaceutical University (approval numbers 2024-02-007 and 2024-09-130).
Neutropenic mouse thigh-infection model
Specific-pathogen-free female ICR mice, aged 6 weeks and weighing 23–27 g, were used in the current study (Hangzhou Medical College, China). The mice were randomly housed in individual cages and acclimatized for 3 days before the experiments. Neutropenia was induced on day 1 and day 4 through intraperitoneal injection of 150 mg kg−1 and 100 mg kg−1 of cyclophosphamide, respectively. A singe colony of C. albicans BNCC 186382, C. albicans BNCC 357785, Cryptococcus neoformans BNCC 225501 or amphotericin-B-resistant C. auris AMR005 was inoculated into 5 ml YPD liquid medium and shaken overnight at 30 °C, 220 rpm. The overnight fungal culture was washed three times with 0.9% sterile saline solution and then diluted to a final concentration of 2 × 107 colony-forming units (CFU) per ml. The diluted fungal suspension (50 µl) was administered by intramuscular injection into both thighs, which provides an inoculum of approximately 1.0 × 106 CFU in each thigh. For the thigh-infection model with the Cryptococcus strain, mice were treated with mandimycin (formulated in 10% DMSO + 10% Tween 80) at a dose of 10 mg kg−1, once daily, 6 h after infection. Amphotericin B, rezafungin, isavuconzole and 5-fluorocytosine were used as controls. For the thigh-infection model with the Candida strain, mice were treated with mandimycin (formulated in 10% DMSO + 10% Tween 80) at doses of 20 mg kg−1, 10 mg kg−1 and 5 mg kg−1 or with the vehicle (10% DMSO + 10% Tween 80) at 2, 10 and 18 h after infection. The mice were euthanized, and their thigh muscles were aseptically removed, weighted, homogenized and enumerated for fungal burden by CFU counts after plating on YPD agar and incubated at 30 °C. All graphical data are presented as individual data points per group and were statistically analysed using GraphPad Prism 9. All animal study procedures were approved by the Animal Ethics Committee of China Pharmaceutical University (approval numbers 2023-08-018, 2023-09-036 and 2023-09-033).
Neutropenic mouse model for disseminated candidiasis
Specific-pathogen-free female ICR mice, aged 6 weeks and weighing 23–27 g, were used for the current study (Hangzhou Medical College, China). The mice were randomly housed in individual cages with four mice per cage and acclimatized for 3 days before the experiments. A singe colony of C. albicans BNCC 186382 was inoculated into 5 ml of YPD liquid medium and shaken overnight at 30 °C, 220 rpm. The overnight fungal culture was washed three times with 0.9% sterile saline solution and then diluted to a final concentration of 2 × 107 CFU ml−1. Subsequently, 50 µl of the diluted fungal suspension was administered by subcutaneous injection into the tail vein, resulting in an inoculum of approximately 1 × 107 CFU in the bloodstream. Six hours after infection, mice were treated with a single dose of 1 mg kg−1, 3 mg kg−1, 5 mg kg−1 or 10 mg kg−1 of mandimycin (formulated in 10% DMSO and 10% Tween 80) through subcutaneous injection. For the oral administration group, mice were orally administered a dose of 10 mg kg−1 of mandimycin formulated in a solution containing 5% DMSO and 10% Tween 80. Twenty-four hours after infection, mice were euthanized, and their kidney and lung tissues were aseptically removed, weighted, homogenized and subjected to fungal burden enumeration by CFU counts after plating on YPD agar and incubated at 30 °C. All graphical data are presented as individual data points per group and were statistically analysed using GraphPad Prism 9. All animal study procedures were approved by Animal Ethics Committee of China Pharmaceutical University (approval numbers 2024-04-009 and 2024-03-019).
Neutropenic mouse model for invasive candidiasis
Specific-pathogen-free female ICR mice, aged 6 weeks and weighing 23–27 g, were used for the current study (Hangzhou Medical College, China). The mice were randomly housed in individual cages with six mice per cage and acclimatized for 3 days before the experiments. To induce immunosuppression, the mice received an intraperitoneal injection of cyclophosphamide (200 mg kg−1) and subcutaneous administration of cortisone acetate (500 mg kg−1) on day −2 and day 3, respectively. To prevent cross-infections, mice were given enrofloxacin orally at a concentration of 50 μg ml−1 in drinking water from day 1 to day 3, followed by subcutaneous injections of ceftazidime (5 μg per dose) from day 0 to day 9. Invasive candidiasis was induced by intravenous injection of 1 × 106 CFU of C. albicans BNCC 186382. Treatment was started 16 h after infection, involving subcutaneous injections of ceftazidime and daily single doses of mandimycin at 1 mg kg−1, 5 mg kg−1, 10 mg kg−1 and 20 mg kg−1 as well as amphotericin B at 10 mg kg−1 for four consecutive days. The mice were monitored for a total of 20 days, and survival rates were plotted using GraphPad Prism 9. All animal study procedures were approved by Animal Ethics Committee of China Pharmaceutical University (approval number 2024-01-014).
Neutropenic mouse skin-infection model
The mouse skin-infection mode was established to evaluate the efficacy of mandimycin in treating fungal infections on the skin47,48,49. An equal number of male and female BALB/c mice weighing 20–22 g were used for the current study (Hangzhou Medical College, China). The mice were randomly housed in individual cages with four mice per cage and acclimatized for 3 days before the experiments. Neutropenia was induced by intraperitoneal injection of 50 mg kg−1 of cyclophosphamide on the third and first days before infection. Subsequently, mice were anaesthetized with 50 mg kg−1 of pentobarbital sodium by intraperitoneal injection and underwent full-thickness skin perforation on the dorsal skin using a biopsy puncher with a diameter of 0.8 cm. A suspension of C. albicans BNCC 186382 (1 × 108 CFU ml−1, 50 μl per mouse) was inoculated into the circular wound, followed by gentle airflow until the skin appeared moist but without excess liquid. One day after infection, the wounds were topically treated with mandimycin (2.5 mg kg−1 or 7.5 mg kg−1), amphotericin B (2.5 mg kg−1 or 7.5 mg kg−1) or vehicle (PBS containing 10% DMSO and 10% Tween 80). The mice that received a wound but no fungal infection served as negative controls and were treated with vehicle only. All compounds were administered once daily for five consecutive days. The wounds were photographed and their sizes were measured on days 1, 5, 9 and 11 after infections. On day 11, fungal counts of wound specimens were recorded, and wound specimens were collected for H&E staining. All animal study procedures were approved by Animal Ethics Committee of China Pharmaceutical University (approval number 2024-02-003).
Neutropenic mouse model for vaginal candidiasis
Female BALB/c mice, weighing 19–21 g, were acclimatized for 3 days before the experiments. The mice were randomly housed in individual cages with four mice per cage and acclimatized for 3 days before the experiments. On day 1, mice were subcutaneously injected with 10 mg kg−1 of oestradiol benzoate, once daily for five consecutive days to induce oestrus50,51. On day 6, mice were intravaginally inoculated with 50 μl of C. albicans BNCC 186382 suspension (1 × 1010 CFU ml−1) using a pipettor and were subsequently positioned upside down for 5 min after vaginal inoculation. After 3 days of consecutive infection, the mice were fed normally ad libitum food for 1 day before receiving a subcutaneous administration of mandimycin (10 mg kg−1), amphotericin B (10 mg kg−1), rezafungin (10 mg kg−1) or isavuconazole (10 mg kg−1) once daily for five consecutive days. Mice infected with C. albicans but not treated with compounds served as the control group and were administered PBS containing 10% DMSO and 10% Tween 80. On the second day after the final injection, vaginal douche was obtained by repeatedly washing the vagina with sterile PBS (20 μl) by pipetting, and the samples were plated on De Man, Rogosa and Sharpe (MRS) agar plates for counting C. albicans colonies. Finally, all animals were anaesthetized with diethyl ether and euthanized to collect the vaginal tissues, which were subsequently fixed with 4% paraformaldehyde. The tissues were dissected and subjected to H&E and periodic acid–Schiff staining for histological analysis. All animal study procedures were approved by Animal Ethics Committee of China Pharmaceutical University (approval number 2024-02-003).
Single-dose pharmacokinetic study of mandimycin
Specific-pathogen-free male Sprague–Dawley rats (180–220 g, 7–8 weeks old, n = 3 per group) were used in the study. The rats were randomly housed in individual cages with three rats per cage and acclimatized for 3 days before the experiments. Rats were administered 25 mg kg−1 of mandimycin through subcutaneous injection. Blood samples (around 0.15 ml) were collected from the jugular vein catheter into tubes containing heparin sodium at 5, 10, 20 and 30 min before dosing and at 1, 2, 4, 6, 8, 12 and 24 h after dosing. After collection, each blood sample was placed on ice and subsequently centrifuged (8,000g, 5 min) to separate plasma. Plasma was then transferred and immediately frozen (at or below −70 °C) until analysis. Mandimycin in rat plasma was analysed using HPLC–ESI–MS/MS on an AB SCIEX Triple Quad 6500 system coupled with a HPLC system equipped with a Quaternary Solvent Manager-R solvent distribution unit and Sample Manager FTN-R auto-sampler. Diazepam was used as an internal standard. Multiple reaction monitoring with positive ion mode was used to quantify the for mass of the analytes at m/z 1,198.30 to m/z 725.20 for mandimycin and m/z 285.00 to m/z 193.00 for the internal standard. Separation of mandimycin and the internal standard was performed through HPLC (Waters ACQUITY C18 column, 1.9 μm, 100 × 2.1 mm). The isocratic mobile phase, consisting of 80% acetonitrile and 20% (v/v) 5 mM ammonium acetate, was delivered at a rate of 0.4 ml min−1 for 3 min through the mass spectrometry electrospray ionization chamber. The concentration in plasma was fitted against time using GraphPad Prism 9. The maximum plasma concentration (Cmax), time to Cmax, apparent elimination half-life, mean residue time, area under the plasma concentration–time curve, clearance and volume of distribution values were estimated by non-compartmental analysis methods using Phoenix WinNonlin 8.3. Bioavailability was calculated by (AUCsubcutaneous/AUCintravenous) × (Doseintravenous/Dosesubcutaneous) × 100%. All animal study procedures were approved by Animal Ethics Committee of China Pharmaceutical University (approval number 2024-02-006).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.