Animals
Mice were maintained on a 12 h:12 h dark-light cycle with access to food and water ad libitum. The temperature was maintained at 22 °C and the humidity between 30% and 70%. All experiments were performed in accordance with National Institutes of Health guidelines and approved by the Harvard University Institutional Animal Care and Use Committee. Experiments were performed in adult female mice to avoid interfering behaviours such as aggression or mating seen in male mice after social isolation. The following mouse lines (strain no.) were obtained from the Jackson Laboratory: BALB/cJ (000651), DBA/2J (000671), C57BL/6J (000664), C3H/HeJ (000659), SWR/J (000689), FVB/NJ (001800), sighted FVB (FVB-Pde6b+, 004828), TRAP2 (also called Fos2A-iCreERT2, 030323), Ai9(RCL-tdT) (007909), Mc4r-2a-Cre (030759), Vglut2-IRES2-FlpO-D (030212), VGAT-2A-FlpO-D (029591), ROSA-DTA (009669) and Nav1.8-Cre (036564). We obtained Pde6brd1 mutant mice in C57BL/6J background from C. Cepko (Harvard Medical School) and Mrgprb4-Cre mice from I. Abdus-Saboor (Columbia University). Trpc2 knockout mice were generated previously in our lab36. The following mouse lines were backcrossed to FVB/NJ mice for (N) generations before experiments: TRAP2/Ai9 (N > 9, activity labelling experiments), Vglut2-Flp and VGAT-Flp (N ≥ 5, neural tracing and optogenetics), Mc4r-Cre (N = 3, calcium imaging and optogenetics), Mrgprb4/DTA, Nav1.8/DTA and Trpc2 (N = 1, behavioural assay). The sample sizes for experiments were chosen on the basis of common practices in animal behaviour experiments.
Behavioural assays
All behavioural experiments were performed during the dark cycle of the animals in a room illuminated by infrared or red light. Mice were habituated in the room for 10–20 min before experiments. For anxiety and stress tests, mice were acclimatized to the testing environment for 1 h before testing to reduce basal stress level.
Social isolation/reunion assay
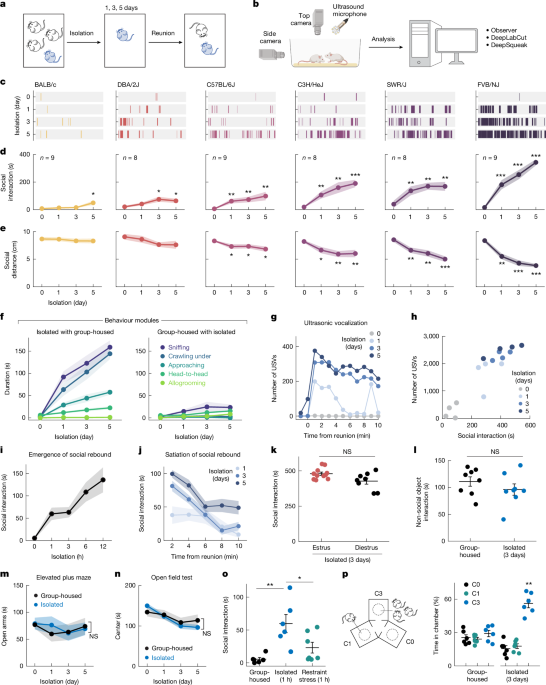
Female sibling mice were group housed (at least three mice) after weaning for at least 1 week (4-week-old mice) before social isolation experiments. Before isolation, two group-housed cagemates were put together in a new cage with fresh bedding for 10 min to measure the baseline social interaction (isolation day 0). One mouse was then isolated in their home cage or in a new cage for 5 days while the other was kept in group. On the first, third and fifth day from the onset of isolation, the isolated mice were reunited transiently with the same group-housed cagemate in a new cage for 10 min. In each reunion session, the isolated mice were first put in the cage, and then its group-housed cagemate was introduced. All behaviours occurring during the reunion period were recorded using a multi-camera surveillance system (GeoVision GV-VMS software and GV-BX4700-3V cameras). Behaviour videos were scored manually using the Observer XT 11 software (Noldus Information Technology) to identify typical social behavioural modules, including approaching (one mouse moves towards another), sniffing (the nose of one mouse comes close to or makes contact with another mouse’s body, usually the anogenital region), crawling under (one mouse crouches down, crawls underneath another mouse’s body and sometimes passes through), head-to-head contact (two mice approach each other and contact each other’s noses) and allogrooming (one mouse grooms another mouse, usually on the head, neck and back regions). Every single event of these behavioural modules was considered as a social interaction bout, and the intervals between social interaction bouts were measured as behavioural latency. The transition probability of one behavioural module occurring after a different module was calculated across all reunion sessions to quantify the unique motor sequences of social interaction during reunion (Extended Data Fig. 1f). The total durations of all the modules within one reunion session were summed up to calculate total social interaction, and the social interactions initiated by the isolated mice were used to indicate social rebounds unless otherwise noted. To demonstrate the satiation process during social rebound, we calculated social interaction in discrete time bins (2 min per bin) during reunion. Social interactions were also measured after 1, 3, 6 and 12 h of isolation or 7 and 10 days of isolation to show the emergence of social rebound or its plateau phase. Custom MATLAB codes and DeepLabCut software package were used to track frame-by-frame positions of two mice during social reunion, and the social distances between two mice were calculated and averaged across frames during one reunion session (Fig. 1e). Because 1 and 3 days of social isolation are sufficient to trigger significant social rebound in FVB/NJ mice, we used these two isolation schedules in our functional manipulation experiments.
USV detection
USVs during social reunion were recorded in a sound isolation box using an ultrasonic microphone (Ultrasound Gate CM16/CMPA; Avisoft Bioacoustics) positioned 30 cm above the floor of the cage and converted into a digital format by an analogue-to-digital converter (Ultrasound Gate 116, Avisoft Bioacoustics) sampling at 500 kHz. The Avisoft Recorder software was used to control the recording and audio files were saved as 16 bit WAV format and later analyzed with DeepSqueak50—a deep learning-based system for detection and analysis of USVs. The built-in mouse call detecting neural network was used to identify USV syllables, and the detected syllables were reviewed manually to correct false labelling. The corrected detection files were then exported as .mat files. Custom MATLAB code was used to extract the timing and duration of each USV. The number of USVs was plotted as a function of different timecourses to show the dynamic changes within and across reunions. As only one microphone was used for recording, we were not able to determine the source of USVs; however, as group-housed female mice emit few USVs, and we detected USVs from isolated mice before cagemates were introduced, we assumed that most of the recorded USVs during reunion were probably generated by the isolated mice. Correlation between USVs and social interaction initiated by the isolated mice were assessed over time.
Non-social object interaction test
To test whether social isolation increases the motivation for general investigation behaviours, we tested behaviour towards a non-social object in group-housed or isolated mice. Before isolation, group-housed mice were presented with a 15 ml centrifuge tube or a rubber toy mouse in a new cage for 10 min to measure baseline behaviour. Mice were then isolated in their home cage or in a new cage for 3 days before a second test with the object. Behaviour during tests was recorded and analyzed. Any contact or climbing behaviours with the object were scored and added up as the time spent interacting with the object.
Social interaction in different phases of oestrous cycle
To test whether social rebound in female mice after social isolation is influenced by the oestrous stage of the animal, we performed reunion assays after 3 days of social isolation and identified the oestrous phase of each tested mouse. Time spent in social interaction was compared between mice in either oestrus or dioestrus phases. Vaginal smears were examined under the microscope, and specific oestrous stages were characterized on the basis of the morphology of vaginal epithelial cells as described previously51. In brief, we collected vaginal cells from female mice with 10 µl of PBS and observed these samples under a light microscope with a ×40 objective to characterize the morphology of cells. During the oestrus phase, vaginal epithelial cells are cornified and appear large and flat, while during the dioestrus phase, these cells are smaller with round shapes51.
Anxiety and stress assays
Mice’s anxiety levels were measured after different durations of social isolation using elevated plus maze and open field tests. The elevated plus maze was on a pedestal 1 m above the ground and consisted of two closed arms (30 × 5 cm2 with 15-cm-high wall) and two open arms (30 × 5 cm2) arranged 90° from each other with a central platform (5 × 5 cm2), all made of black acrylic board. Mice were placed on the central platform and allowed to freely explore the maze for 5 min while the behaviour was recorded. All behaviour videos were scored manually with Noldus Observer software to measure the total time spent in the open and closed arms. In the open field test, individual mice were placed in a 42 × 42 × 42 cm3 arena composed of black acrylic board and allowed free exploration for 10 min. The behaviour was recorded and analyzed to measure the total time spent in centre (24 × 24 cm2) and peripheral zones. To induce physical restraint stress, individual mice were placed into 50 ml conical tubes with ventilation holes for 1 h and then reunited with one of their cagemates in a new cage to monitor the behaviours with the same settings as described for social reunion assays.
Social preference tests
To examine the preference of the tested mice for a group versus a single mouse, we built a new arena with three cubic chambers (25 × 25 × 25 cm3) joined by a triangular central zone to allow for unbiased entry into any chamber (Fig. 1p). Each chamber contained an inverted wire cup. The cup was empty in the C0 chamber, contained one cagemate in the C1 chamber and contained three cagemates in C3 chamber. The locations of the three types of chamber were assigned randomly across animals. Group-housed or isolated mice were first introduced into the central zone at the beginning of the test and allowed to explore the arena freely for 10 min. Behaviours were recorded and time spent in each chamber was scored manually. In the preference test between unfamiliar mice versus cagemates (Extended Data Fig. 1m), a standard three-chamber task was used in which the arena consisted of two choice chambers on two sides and a central zone that allowed for unbiased entry into any chamber. Each of the two choice chambers contained an inverted wire cup with either a familiar mouse (cagemate) or a stranger from the same strain. The tested mice (group-housed or isolated) were first introduced into the central zone at the beginning of the test and allowed to explore the arena freely for 10 min. Behaviour was recorded and time spent in each chamber was scored. The same arena and experimental procedure were used in the strain preference test, in which the two choice chambers contained mice from either the same or different strains.
Sensory modality screening
To investigate the contribution of different sensory modality to the emergence of social rebound after isolation, we designed a home-cage divider experiment in which a plastic divider with laser-cut thin slots/openings was placed in the diagonal of home cage to subdivide a group of three mice, such that one mouse was placed into one side and the other two together on the other side. Food, water and nesting materials were provided equally on both sides. The divider prevented the separated mice from physically interacting with each other but allowed exchange of auditory and olfactory information. After 3 days of separation, the singly divided mouse was reunited with one of the mice from the other side in a new cage for 10 min. The occurring behaviours were recorded and scored manually. The time spent in social interaction was compared with the social rebound after 3 days of isolation in the singly housed condition. To examine the potential contribution of pheromone sensing to social rebound, we assessed the behaviour of Trpc2−/− mice, which are impaired in vomeronasal pheromone sensing. Trpc2−/− mice were first crossed to FVB/NJ strain for one generation, and the resulting Trpc2+/− mice were used to cross to each other to generate wild-type (Trpc2+/+) and mutant (Trpc2−/−) mice for the experiments. We measured the rebound social interaction after 3 days of isolation and analyzed the satiation process in wild-type and mutant mice.
Gentle touch preference test
To test the preference of gentle touch before and after social isolation, we designed a free choice task for mice to interact with either a naked plastic tunnel (10-cm long, provided by Harvard Biological Research Infrastructure) or a tunnel lined inside with a layer of soft plush towel (bought from Amazon), referred to as cloth tunnel. All materials were autoclaved before use and replaced between animals to avoid odour contamination. During the test, group-housed or 1-day isolated mice were placed in a new cage with one naked tunnel and one cloth tunnel. Mice were allowed to explore freely and go through either tunnel for 15 min. Behaviours were recorded, and the crossing events were scored. The touch preference indexes were measured in terms of the number or the duration of crossings through the cloth tunnel divided by all crossings through both tunnels.
Touch manipulation assays
To acutely reduce tactile sensitivity during social reunion, isolated mice received an intraperitoneal injection of isoguvacine37 (20 mg kg−1)—a peripherally restricted GABAAR agonist—60 min before social reunion. PBS was injected in a different batch of isolated mice, which served as a control group. To examine the contribution of tactile sensation to the emergence of social need, we genetically ablated somatosensory neurons marked by Mrgprb4 or Nav1.8, which are thought to mediate social touch in mice39,40,41. We first separately backcrossed Mrgprb4-Cre (B4-Cre), Nav1.8-Cre and Cre-dependent DTA mouse lines (all in C57BL/6J background) to FVB/NJ strain for one generation and then crossed the resulting F1s (F1(B4-Cre/FVB), F1(Nav1.8-Cre/FVB) and F1(DTA/FVB)) to ablate Mrgprb4-lineage neurons (B4/DTA) or Nav1.8+ neurons (Nav1.8/DTA). We measured social interaction in B4/DTA and Nav1.8/DTA mice after 0 or 3 days of isolation and compared these results with the social rebound measured in wild-type mice from FVB/NJ × C57BL/6J cross. In the acute touch rescue experiments (Fig. 5h), faux-fur-lined tunnels were used to provide comfort-touch stimulation as an enhanced version of cloth tunnel described in ‘Gentle touch preference test’. Mice were habituated to the faux-fur-lined tunnels by continuously going through two of these tunnels that were alternatingly connected by experimenter before social isolation. Mice were then isolated for 24 h and reunited with one cagemate both before and after gentle touch stimulation. Specifically, a 5-min reunion assay was first performed to measure the baseline social interaction, and then the mice went across faux-fur-lined tunnels 30 times the same way as in habituation. The 30 crossings typically took around 10 min for both cloth tunnel and naked control tunnel. A second reunion assay was then conducted to measure the change of social motivation compared with the first reunion. Naked plastic tunnels were used in another batch of animals as a negative control. In the chronic touch rescue experiment (Extended Data Fig. 10e), isolated mice were co-housed with either a cloth tunnel or a naked tunnel described in ‘Gentle touch preference test’ for 24 h. A social reunion assay was then carried out to measure and compare social interaction after different co-housing conditions.
Microendoscopy calcium imaging
To examine real-time neuronal activity with single-cell resolution in the MPN, we performed microendoscopy calcium imaging in FVB/NJ (n = 6), C57BL/6J (n = 6) and Mc4r-Cre/FVB mice (n = 3). The Mc4r-Cre mouse line, used for labelling MPNIsolation neurons, was backcrossed for at least three generations to FVB/NJ strain before experiments. All imaging experiments were performed during the dark cycle of the animals in a chamber illuminated by infrared light.
Virus injection and GRIN lens implantation
For pan-neuronal activity imaging, 400 nl of AAV1-Syn-GCaMP6s (Addgene, catalogue no. 100843-AAV1) was injected unilaterally into the MPN of FVB/NJ or C57BL/6J mice, at anterior-posterior 0, medial-lateral 0.3 and dorsal-ventral −4.8 (Paxinos and Franklin atlas). To image MPNMc4r+ neurons, 400 nl of AAV1-Syn-Flex-GCaMP6s (Addgene, catalogue no. 100845-AAV1) was injected unilaterally into the MPN of Mc4r-Cre/FVB mice using the above coordinates. As the brain anatomy of FVB/NJ strain differs slightly from the Paxinos and Franklin brain atlas, we adjusted the anterior-posterior coordinate 0.4–0.5 mm towards the rostral side to target the MPN in FVB/NJ mice. At 30 min after viral injection, a 25-gauge blunt needle (SAI infusion technologies, VWR, catalogue no. 89134-146) was slowly inserted (1 mm per 5 min) into the brain targeting anterior-posterior 0, medial-lateral 0.3 and dorsal-ventral −4.7 (Paxinos and Franklin atlas) to create a tract. The needle then was withdrawn slowly, and a GRIN lens (Inscopix, 0.6 × 7.3 mm) was inserted slowly (1 mm per 10 min) into the tract formed by the needle and targeted at 100 µm below the end of the needle tract. The lens assembly with the baseplate was secured on the skull with dental cement (Parkell), and a titanium headplate was attached at the base of the lens assembly with dental cement to restrict the animal’s head for attaching to the microendoscope. The positions of all implanted GRIN lenses were assessed using post hoc histology and only the imaging data from correctly targeted lens were used for further analysis. Mice were housed individually after surgery for 1 week and then co-housed with one or two cagemate(s) for another 3–4 weeks to allow for the expression of GCaMP and clearing of the imaging window.
Calcium imaging and behavioural assays
Optimal imaging settings (focal and excitation parameters) were determined on the first day of the imaging experiments. The implanted mouse was restrained transiently and the microendoscope (nVista3, Inscopix) was attached onto the lens assembly on the head of the mouse. The focusing plane of the microendoscope was adjusted carefully over the entire working distance to choose an imaging plane with most neurons and sharp image. The field of view was cropped to the region encompassing the entire lens field of view. The illumination power (roughly 10% of the maximum power) and the sensor gain (roughly 10–20% of the maximum gain) were chosen to have a strong relative increase in fluorescence signal, but not saturated. These imaging settings were saved for each animal and used subsequently for the same animal across sessions. We used inbuilt acquisition software from Inscopix to acquire images at 10 Hz. Before formal data acquisition, mice were habituated to the recording setup and environment two to three times. An Arduino microcontroller is programmed to send transistor–transistor logic (TTL) signals to synchronize the data acquisitions of calcium signals (DAQ, Inscopix), animal behaviour recoding (BFS-U3-31S4M-C, FLIR Blackfly S camera) and USV recoding (Avisoft Bioacoustics). In the social reunion assays, implanted mice were isolated for 0, 1, 3 or 5 days before imaging. During the imaging sessions, there was an initial 5- or 10-min baseline period during which the mouse was kept alone. Following this, a cagemate was introduced into the cage, and social interaction between the mice was allowed for 5 or 10 min. Subsequently, the cagemate was removed, and the mouse was kept alone again for another 5 or 10 min. To monitor the neuronal activity during the initial 6 h of social isolation but avoid GCaMP signal photobleaching, we performed 15 min of imaging every hour for 6 h with the microendoscope connected during the whole 6-h procedure. This allowed us to track the same neurons across 6 h of isolation. We provided food and hydrogel in the cage to ensure that the mice did not experience hunger or thirst. At the end of the 6-h imaging, we performed a 5-min reunion assay to identify neurons with significantly modulated activity. To examine the activity specificity, we performed a series of control experiments in which a 3-day isolated mouse encountered (for 10 min) one control stimulus, including food pellets, a C57 stranger female, a pup and a castrated male. Then we removed the control stimulus and left the mouse alone for a 15-min interval, followed by a 5-min reunion assay to identify the significantly modulated neurons. During the whole procedure, the microendoscope was connected to the animal to track the same neurons across different events. Distinct stimulation was presented in separated imaging sessions with a random order to avoid the influence across stimulations. We used a castrated male to provide social companion and avoid mating or aggressive behaviours. Before imaging with food pellets, the mouse was food restricted overnight. Tail suspension was performed by suspending the mouse manually by the tail for up to 2 min and usually after reunion to avoid its stressful influence on social behaviour. In the tunnel crossing experiment, the implanted mouse was isolated for 3 days before imaging. During the imaging session, there was a 5-min baseline period followed by 15 min of free tunnel crossing during which two faux-fur-lined tunnels (10 cm long, 7.5 cm in diameter, the fur around 1.5 cm long) were introduced into the cage to maximize crossing behaviours. The top of the tunnel was removed creating a slot (4 cm) to allow the implanted mice to run through with the attached wire. The tunnels were subsequently removed from the cage and the mouse was kept alone for 30 min. Finally, we performed a 10-min imaging with 5 min of baseline and 5 min of reunion to identify MPNIsolation and MPNReunion neurons.
Image processing and calcium signal extraction
The images acquired were processed in two steps. In the first step, we processed the raw image data using the Inscopix data processing software (IDPS v.1.8.0.3519). The images were imported in the proprietary Inscopix format to IDPS. The images were spatially downsampled by a factor of four to reduce the file sizes for subsequent steps without losing quality, followed by a spatial bandpass filter in the frequency band of 0.005 to 0.5 per pixel. The images were next subjected to motion correction using IDPS and the resulting images were saved as a tiff image stack. The second step used standard MATLAB scripts from the CNMFe52 database (https://github.com/zhoupc/CNMF_E). Calcium traces were extracted and deconvolved using CNMFe pipeline with the following parameters: patch_par = [2,2], gSig = 3, gSiz = 13, ring_radius = 9, min_corr = 0.8, min_pnr = 8, deconvolution: foopsi with the ar1 model52. The spatial and temporal components of every extracted unit were inspected carefully manually (SNR, PNR, size, motion artifacts, decay kinetics, and so on) and outliers (obvious deviations from the normal distribution) were discarded. Cells with elongated and thin shapes were removed using custom MATLAB codes.
Single-neuron and population activity analysis
All analysis based on extracted calcium traces were performed using custom MATLAB scripts. To categorize the pan-neuronal calcium activity patterns at single-neuron level during social reunion, reunion induced responses were calibrated for each neuron using ROC analysis as described previously28,53. ROC curves were calculated by comparing the distribution of raw calcium responses during baseline (isolation, 300 s) and during reunion (300 s) (Extended Data Fig. 2a,b). A series of thresholds moving from the minimum calcium response to the maximum were set to evaluate the binary separation of the calcium activity from baseline versus reunion. For each threshold, we calculated the probability that the baseline activity was greater than the threshold (falsely categorized as reunion activity, that is, false positive rate) and the probability that reunion activity was greater than the threshold (correctly categorized as reunion activity, that is, true positive rate). An ROC curve was then plotted by graphing the true positive rate against the false positive rate for all thresholds. The area under the ROC curve (AUC) was used to quantify the response strength of each neuron. AUC values more than 0.5 indicate increased response compared with baseline, whereas AUC values less than 0.5 indicate decreased response. The significance of the responses was determined by comparing AUC from real data and randomly shuffled data. Specifically, we randomly shuffled the calcium activity in each time bin (1 s) between baseline and reunion period 1,000 times and calculated the AUC values in each shuffle. Neurons with the AUC value >0.7 or <0.3 and exceeded the 95th percentile of the AUC distribution from shuffled data, were considered significantly tuned by reunion, among which the inhibited neurons are referred to as MPNIsolation neurons and the activated neurons are referred to as MPNReunion neurons. We used the AUC values to infer the activation strength of MPNReunion neurons during reunion and (1 − AUC) values to infer the (relative) activation strength of MPNIsolation neurons during isolation (baseline) period. In the imaging experiments during the initial 6 h of isolation, neurons with the activity that met the above standard for at least 1 h were identified as MPNIsolation neurons. Similar ROC analyses were applied to the neuronal responses during other imaging experiments. The total duration of activation or inhibition for each neuron during reunion was calculated by counting the time bins where calcium activity was modulated significantly, either above 2 s.d. from the baseline mean or below 30% of the baseline mean. The duration values from each neuron were averaged to obtain the mean for the animal (Extended Data Fig. 2h). The persistent activation or inhibition for each neuron was quantified as the longest duration where the calcium activity in all time bins was modulated significantly, and the duration values from each neuron were averaged to obtain the mean for the animal (Extended Data Fig. 2i). To visualize the population activity during social reunion assay, we performed PCA with significantly tuned neurons from all imaged mice with the same isolation schedule and projected their activity (z scores) onto the first three principal components to visualize the neural trajectory across time (bin size, 5 s; Fig. 2g). To quantify state changes, the Euclidean distances between each point in the trajectory (time bin, 1 s) and the mean of the baseline were calculated using the minimum number of principal components that explain 95% of the variance. This same procedure was repeated 100 times by selecting 50% of the neurons randomly from the imaging sessions to calculate the mean and s.d. (Fig. 2h). To describe how the isolation state is re-established during the re-isolation period, when the partner mouse was removed after a brief reunion, the latency for the PCA distances to return to the initial isolation (baseline) state and the durations after return were calculated using the upper bound of the 95% confidence interval of the baseline as a threshold (Extended Data Fig. 3a). In the population activity decoding analysis of isolation versus reunion states, we trained support vector classifiers using 80% of the data that were selected randomly from each state and tested the accuracy of the classifiers with the remaining 20% data (Extended Data Fig. 3b). To identify the neurons with significant ramp-up or ramp-down activity during the initial 6 h of isolation, calcium activity averaged in 5-min bins was used to run a linear correlation test against isolation durations using the ‘corrcoef’ function in MATLAB, and the neurons with P values < 0.05 were considered to have significant ramp-up or ramp-down activity (Extended Data Fig. 2j).
In cell-type targeted imaging experiments, we monitored the calcium activity of MPN Mc4r+ neurons from three mice in FVB background, identified significantly inhibited neurons during reunion and examined their activity during other behavioural paradigms, including eating and tail suspension. To identify the same neurons across imaging sessions, we aligned the fields of view from different imaging experiments. If the overlaps between neuron pairs were higher than 70%, these neurons were considered the same. Alternatively, for the experiments performed on the same day (Fig. 5i and Extended Data Fig. 3c–h), the raw imaging data from two sessions in the same field of view were concatenated together to extract calcium traces.
Cell-type identification
TRAP induction and activity labelling
To specifically label neurons activated during social isolation, we took the advantage of the TRAP2/Ai9 mouse line16 that enables the labelling of activated neurons over a 3–6 h time window, which better integrates neuronal activity representing a persistent state compared with Fos in situ hybridization. The TRAP2/Ai9 line was backcrossed to FVB/NJ strain for more than nine generations before use. The isolation-activated neurons were ‘TRAPed’ in the following steps: 10 mg of 4-OHT (Sigma Aldrich, catalogue no. H6278-10mg) was added with 500 μl ethanol and shaken at room temperature until the powder was completely dissolved (20 mg ml−1). The resolved 4-OHT solution was then mixed with corn oil in 1:2 volume ratio and vortexed to fully mixed to extract the 4-OHT in the oil. The resulting solution was vacuum centrifuged for 1–1.5 h until the upper layer of ethanol was evaporated. The final 10 mg ml−1 4-OHT solution was injected intraperitoneally into mice immediately after preparation at a dose of 50 mg kg−1. To label (‘TRAP’) the cells activated during social isolation, mice were isolated for 3 days and injected with the 4-OHT solution in the dark phase of the third isolation day. After at least 7 days, the mice were euthanized for in situ hybridization experiments. To assess the activity of PBN during isolation, we performed TRAP inductions after 1 or 3 days of isolation in different cohorts of animals. To label the neurons that are activated during social reunion, FVB/NJ mice were isolated for 3 days and reunited with one of its cagemate. At 30–40 min after the onset of the reunion, mice were euthanized for in situ hybridization experiments. The reunited mice were kept with cagemate from reunion assay until euthanasia to prevent the activation of isolation-related neurons. To assess the activity of PBN during reunion, 4-OHT was injected 1 h after reunion and the reunited mice were kept in group for at least 24 h to allow tamoxifen to be fully metabolized.
In situ hybridization
RNAscope v.2 kit (Advanced Cell Diagnostics (ACD)) was used to perform double-label and triple-label fluorescence in situ hybridization according to the manufacturer’s instructions. Probes for tdTomato and Fos were used to visualize activated neurons labelled with TRAP method or acute behavioural assays, respectively. Probes for marker genes of specific cell types were selected from previous single-cell RNA sequencing and functional studies. All probes were made by ACD. Animals were euthanized after specific behavioural assays and the brains were dissected. Freshly frozen brains were sectioned using a cryostat at 16 μm and stored at −80 °C. On the day of the fluorescence in situ hybridization experiment, slides were thawed and fixed in 4% paraformaldehyde (PFA) for 15 min followed by dehydration in 50%, 75% and 100% ethanol at room temperature. Tissue samples were processed using 3% hydrogen peroxide (VWR) for 10 min and permeabilized for 25 min using Protease IV (ACD). For each RNAscope experiment, C2 and C3 probes were diluted in C1 probe solution (1:50), heated to 40 °C for 10 min and applied to slides that were placed in ACD HybEZ II oven at 40 °C for 2 h. Tissue samples were then processed as suggested by the RNAscope v.2 protocol (ACD). Slides were imaged at ×10 on an Axioscan 7 using Zen Blue v.3.5 software (Zeiss). The number of cells marked by specific and overlapping genes was measured using QuPath v.0.3.2.
Neural circuit tracing
Anterograde tracing
Anterograde tracing experiments were performed in TRAP2/Vglut2-Flp mice and TRAP2/Vgat-Flp mice both in FVB/NJ background. TRAP2, Vglut2-Flp and Vgat-Flp lines were backcrossed separately to FVB/NJ strain for more than five generations and the offspring from these lines were crossed to generate TRAP2/Vglut2-Flp/FVB mice for tracing experiments of MPNIsolation neurons and TRAP2/Vgat-Flp/FVB mice for tracing experiments of MPNReunion neurons. All surgeries were performed under aseptic conditions in animals anaesthetized with 100 mg kg−1 ketamine and 10 mg kg−1 xylazine by intraperitoneal injection. Using a programmable nano-injector (Nanoject III, Drummond), 150–200 nl of Cre- and Flp-dependent virus AAV8-hSyn-Con/Fon-EYFP (Addgene, catalogue no. 55650-AAV8) was injected unilaterally into the MPN (anterior-posterior 0, medial-lateral 0.3 and dorsal-ventral −5, Paxinos and Franklin atlas). We adjusted the anterior-posterior coordinate 0.4–0.5 mm towards the rostral side to match the anatomy of the FVB/NJ brain. After surgery, injected mice were housed singly to recover for 1 week and then put together with former cagemates for another week before TRAP induction (see details in ‘TRAP induction and activity labelling’). To visualize the projections of MPNIsolation neurons, TRAP2/Vglut2-Flp/FVB mice were isolated for 3 days and injected with 4-OHT. To visualize the projections of MPNReunion neurons, TRAP2/Vglut2-Flp/FVB mice were isolated for 3 days and injected with 4-OHT 1 h after reunion. After 2 weeks of viral fluorophore expression, animals were perfused transcardially with PBS followed by 4% PFA in PBS. Brains were dissected and post-fixed in 4% PFA overnight. After embedding in 4% low-melting point agarose (Promega, catalogue no. V2111) in PBS, 50-μm coronal sections were cut through the whole brain on a vibratome (Leica) and mounted on slides (VWR, catalogue no. 48311-703) with DAPI-containing mounting medium (Vector Laboratories, catalogue no. H-1200). The brain sections were imaged at ×10 magnification using AxioScan 7 and Zen Blue v.3.5 software (Zeiss). For quantification of projection density, the average pixel intensity in a target region containing EYFP signals was calculated, and the background was subtracted (Zen Blue software, Zeiss). Because injections were unilateral and no labelling was observed in most cases contralaterally, the equivalent region on the contralateral hemisphere was chosen for background subtraction; in cases where contralateral EYFP were present, an adjacent unlabelled region was chosen. The relative density value for each projection region was calculated as the ratio between background-corrected intensities in each region divided by the sum across all the target regions.
Monosynaptic retrograde tracing
Monosynaptic retrograde tracing experiments were performed with an intersectional strategy in TRAP2/Vglut2-Flp/FVB mice and TRAP2/Vgat-Flp/FVB mice (see details in ‘Anterograde tracing’). We first injected 150–200 nl of a 1:1 mixture of two Cre- and Flp-dependent viruses, AAV8-nEF-Con/Fon-TVA-mCherry (Stanford GVVC-AAV-197) and AAV8‐Ef1a‐Con/Fon-oG (Stanford GVVC-AAV-198) unilaterally into the MPN with the same coordinates as in ‘Anterograde tracing’. The injected mice were housed singly for 1 week and reunited with former cagemates for another week before TRAP induction (see details in ‘TRAP induction and activity labelling’). TRAP2/Vglut2-Flp/FVB mice were isolated for 3 days and injected with 4-OHT to allow viral expression in MPNIsolation neurons; TRAP2/Vgat-Flp/FVB mice were isolated for 3 days and injected with 4-OHT 1 h after reunion to enable viral expression in MPNReunion neurons. Two weeks later, 200 nl of G-deleted rabies virus (EnvA-ΔG-rabies-eGFP, Janelia Viral Tools Facility) was injected into the MPN. Seven days later, mice were euthanized, and the brains were dissected, sectioned and imaged with the same procedure as in ‘Anterograde tracing’. Relative input strength was quantified as follows. First the representative sections of input regions were selected and GFP+ cells (presynaptic cells) were counted. The local presynaptic cells in the MPN were estimated by counting GFP+ and mCherry− neurons. The relative input density was calculated as the ratio between number of presynaptic cells in each input region divided by the sum across all calculated regions in each brain. To identify the input cell types of MPNIsolation neurons, a different cohort of mice were processed for in situ hybridization (see details in ‘In situ hybridization’). Probes for the GFP gene, marker genes or immediate early genes were used to examine the presynaptic cell types.
Optogenetics
Virus injection and fibre implantation
TRAP2/Vglut2-Flp mice and TRAP2/Vgat-Flp mice were injected bilaterally with 200 nl of AAV8-hSyn Con/Fon-hChR2(H134R)-EYFP for optogenetic activation (Addgene catalogue no. 55645) or AAV8-nEF-Con/Fon-iC++-EYFP for optogenetic inhibition (Addgene, catalogue no. 137155) into the MPN (anterior-posterior 0, medial-lateral ±0.3 and dorsal-ventral −5, Paxinos and Franklin atlas) and in the same surgery a dual fibre-optic cannula (200/250-0.66_GS0.6/0.8_FLT, Doric Lenses) was implanted 200 µm above the injection site for MPN cell body manipulation or above the Arc (anterior-posterior −1.6, medial-lateral ±0.3 and dorsal-ventral −5.3) or the LHb (anterior-posterior −1.6, medial-lateral ±0.3 and dorsal-ventral −2.2) for projecting axon terminal manipulation. To manipulate MPN Mc4r+ neurons, 200 nl of AAV-EF1a-DIO-hChR2(H134R)-EYFP (UNC Vector Core) or AAV-EF1a-DIO-iC++-EYFP (UNC Vector Core) was bilaterally injected into the MPN, and the dual fibre-optic cannula was implanted. Mice were recovered for 2 weeks before Isolation-TRAP and Reunion-TRAP induction (see details in ‘TRAP induction and activity labelling’). Mice were tested 3–5 weeks after TRAP induction to allow for efficient expression of ChR2 or iC++.
Optogenetic manipulations
On testing days, the implanted optic fibres were attached through a patch cord (SBP(2)_200/230/900-0.57_FCM-GS0.6/0.8, Doric Lenses) and a rotary joint (FRJ_1 × 1_FC-FC, Doric Lenses) to a 460-nm blue LED module (Prizmatix) for optogenetic activation or inhibition. An Arduino microcontroller is programmed to send TTL signals to a light-emitting diode (LED) module to control the stimulation patterns. Pilot experiments were conducted to test and determine the proper ranges of LED power in different manipulation experiments. After connected to the patch cord, mice were transferred to a new cage with the same setup in social reunion assay and allowed to habituate for 5–10 min. To activate Vglut2+/Isolation-TRAPed neurons or Mc4r+ neurons, the LED was on for 1 s (20 Hz, 10 ms pulses, 6–8 mW at patch cord tip) and off for 3 s, repeatedly. To activate Vgat+/Reunion-TRAPed neurons, the LED was on for 40 s (20 Hz, 20 ms pulses, 6–8 mW) and off for 20 s, repeatedly. In the real-time place preference/avoidance tests, the LED was turned on (20 Hz, 6–8 mW) during the period when mice entered the LED-on chamber assigned randomly in each session. To inhibit Vglut2+/Isolation-TRAPed neurons or Mc4r+ neurons or Vgat+/Reunion-TRAPed neurons, the LED was on for 3 min (constant on, 3–4 mW) and off for 20 s, repeatedly. In the social interaction/reunion tests and three-chamber social preference tests, the patterned LED was applied for 10 min and the behaviours during the entire period were recorded and analyzed as the LED-on performance. In LED-off sessions, the same cohorts of animals were tested without LED stimulation. To minimize potential influence from previous experiments, we usually performed LED-off sessions before LED-on sessions with at least 3–5 days of interval between two different paradigms. Two to three batches of animals were prepared to repeat the results with a randomly assigned order of paradigms to control possible cross-paradigm influence. In the pre-reunion activation experiments, the patterned LED was applied for 5, 10 or 20 min when the animal was alone, followed by a 10-min reunion test with LED off. In the pre-reunion inhibition experiment, the LED was applied for 10 min before reunion. The real-time place preference/avoidance tests lasted for 10 min, and the patterned LED was applied when mice entered the LED-on chamber. For axon terminal manipulation experiments, we used the same protocols as in soma stimulation experiments described above. When testing the stimulation effects on food intake, mice were fasted overnight before experiments. Two food pellets were placed on the two sides of the cage and contacting one pellet triggered the LED on for 10 s whereas contacting the other pellet did not trigger stimulation treated as off controls. Weight reduction of each pellet at the end of a 10-min test was measured as the amount of food intake.
Assessment of oxytocin system
Chemogenetic manipulations of oxytocin neurons
Stereotaxic surgery was used to bilaterally inject 300 nl of AAV8-hSyn-DIO-hM3D(Gq)-mCherry or AAV8-hSyn-DIO-hM4D(Gi)-mChery into the PVN of Oxt-Cre mice using the coordinates medial-lateral ±0.3, anterior-posterior −0.55, dorsal-ventral −4.9. Three weeks after viral injection, mice were used for behavioural tests. In the activation experiments, mice were isolated for 24 h and received intraperitoneal injections of CNO (0.5 mg kg−1) twice: once at the time of isolation and again 9 h before reunion. In the inhibition experiments, mice were isolated for 3 days and received injections of CNO four times: at the time of isolation and once each day thereafter. The final injection was administered 6–9 h before reunion to ensure that the manipulation influenced only the isolation period. Behaviours during reunion were recorded and analyzed as described above.
Oxytocin receptor antagonist
Mice were isolated for 24 h and received intraperitoneal injections of the oxytocin receptor antagonist (OTR-A), L-368,899 hydrochloride (5 mg kg−1) or saline twice as previously described18 first at time of isolation and again 9 h before reunion. Behaviours during the reunion period were recorded and analyzed as described above.
Assessment of dopamine release
Fibre photometry
All surgeries were performed under aseptic conditions with animals anesthetized with isoflurane (1–2% at 0.5–1.0 l min−1). Analgesia was administered pre- (buprenorphine, 0.1 mg kg−1, intraperitoneal) and post-operatively (ketoprofen, 5 mg kg−1, intraperitoneal). We used the following coordinates to target the NAc: anterior-posterior 1.45–1.78, medial-lateral 1.0–1.4 and dorsal-ventral −3.6 to −4.1 from dura. To express dopamine sensor GRABDA2m34, we injected 300 nl of mixed (3:1) virus solution: AAV9-Syn-GRABDA2m (Vigene Bioscience) and AAV5-CAG-tdTomato (UNC Vector Core) unilaterally into the NAc. We then implanted an optic fibre (400 µm diameter, Doric Lenses) slightly above the virus injection site. The implanted mice were housed singly for 1 week to recover and co-housed with previous cagemates for another week before photometry recording. Photometry recording was performed as previously reported54,55. Before recording, we connected a magnetic patch cord (400 µm diameter, numerical aperture 0.48, 3 m long, SMA-SMC, Doric Lenses) to the optical fibre implanted on the head of the animal and the animal was allowed to habituate in a new cage for 10–15 min. Once the recording started, the patch cord simultaneously delivered excitation light at different wavelength (473 nm, Laserglow Technologies; 561 nm, Opto Engine LLC) and collected fluorescence emissions from dopamine sensor and tdTomato (used for motion correction). The emitted light was then filtered using a 493/574 nm beam splitter (Semrock), followed by a 500 ± 20 nm (Chroma) and 661 ± 20 nm (Semrock) bandpass filter, and collected by a photodetector (FDS10 × 10 silicone photodiode, Thorlabs) connected to a current preamplifier (SR570, Stanford Research Systems). This preamplifier outputs a voltage signal that was collected by a data acquisition board (NIDAQ, National Instruments) and custom software written in Labview (National Instruments). Lasers were turned on at least 30 min before recording to allow them to stabilize. Before each recording session, laser power and amplifier settings were adjusted individually for each mouse. The photometry recording and behaviour video acquisition were synchronized using a common TTL input to trigger infrared light pulses (once every 10 s) that were recorded in the behaviour videos. The implanted mice were isolated for 3 days before recording. The recording session lasted for 10 min, including a 5-min baseline period where the animal was kept alone, followed by a 5-min social reunion period where a previous cagemate was introduced.
Fibre photometry data analysis
The tdTomato signal was subtracted from dopamine sensor signal to correct the motion artifacts. The corrected signal was then z scored using the mean and s.d. from a 30-s baseline period before reunion. Individual traces from different mice were aligned at the reunion time point and averaged across animals. The behaviours during recording sessions were scored manually and synchronized with photometry signals by common TTL pulses (‘Fibre photometry recording’).
Dopamine transporter inhibitor infusion
The infusion cannula was prepared according to a previously published protocol56. The cannula was implanted to the NAc bilaterally (anterior-posterior 1.45, medial-lateral 1.4 and dorsal-ventral −3.1 from dura for the guide cannula so that the plug tip/infusion needle was located at dorsal-ventral −4.1) using adhesive cement (C&B Metabond, Parkell) and a small amount of rapid-curing epoxy (Devcon, catalogue no. A00254) together with headplate. The animal was headfixed briefly to connect the infusion needle and released in the home cage. Either dopamine transporter inhibitor (5 mg ml−1; GBR12909, D052, Sigma Aldrich) dissolved in distilled water with 5% dimethyl sulfoxide (Thermo Fisher Scientific, catalogue no. 20688) or vehicle (distilled water with 5% dimethyl sulfoxide) was infused (300 nl per hemisphere with 200 nl min−1 flow rate) with a syringe pump (SP2001, World Precision Instruments) in the home cage. The mice were moved to the reunion cage 5 min after completing infusion and waited for another 5 min before reunion. Behaviours during the reunion period were recorded and analyzed as described above.
Statistics and reproducibility
Data were processed and analyzed using MATLAB and GraphPad Prism v.9. The sample sizes for measuring social rebound in different mouse strains were determined by a MATLAB command ‘sampsizepwr’, which calculates the required cohort sizes to achieve a specified power level (0.8) necessary to confidently detect significant social rebound. In other experiments, the sample sizes were chosen on the basis of common practices in animal behaviour experiments. Individual data points were plotted wherever possible. Error bars and shaded areas in the graphs indicate the mean ± s.e.m. unless otherwise noted. All data were analyzed with two-tailed non-parametric tests unless otherwise noted. In the experiments with paired samples, we used the Wilcoxon matched-pairs signed-rank test or Friedman test. In the experiments with non-paired samples, we used the Mann–Whitney U-test or Kruskal–Wallis test. P values were corrected for multiple comparisons when necessary. Statistical significance is indicted by *P < 0.05; **P < 0.01; ***P < 0.001; NS, no statistical significance. Statistical details are given in the respective figure legends. All behavioural, imaging, in situ hybridization, optogenetics and tracing experiments were replicated in several batches of animals independently with similar results. Experiments were randomized whenever possible. Experimenters were blind to the mouse identity in Mrgprb4-lineage neuron ablation experiments.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.